
Sulfitobacter brevis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Sulfitobacter
Average proteome isoelectric point is 6.22
Get precalculated fractions of proteins
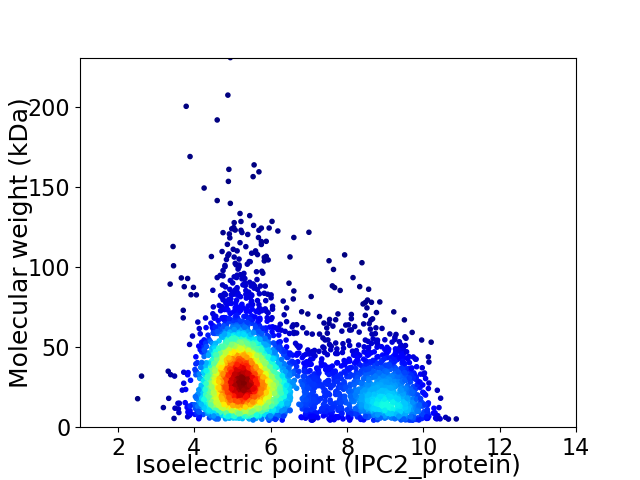
Virtual 2D-PAGE plot for 4154 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
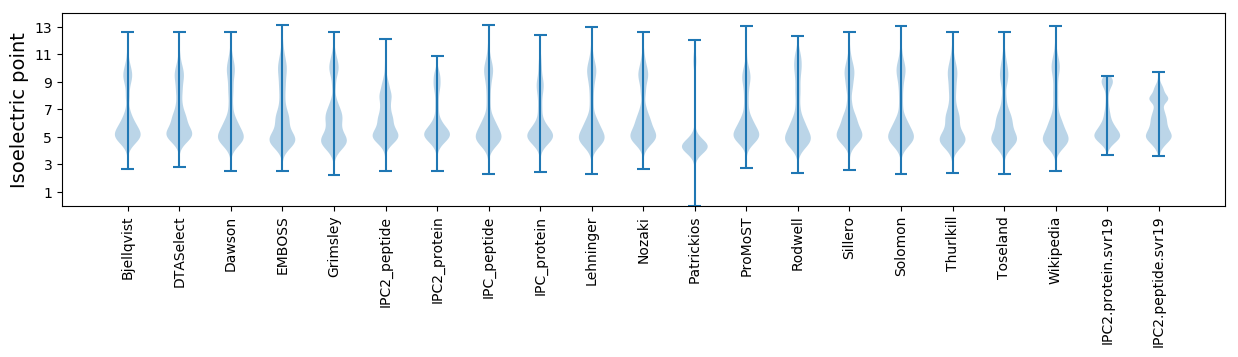
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I1TNU5|A0A1I1TNU5_9RHOB Uncharacterized protein OS=Sulfitobacter brevis OX=74348 GN=SAMN04488523_101414 PE=4 SV=1
MM1 pKa = 7.51TSTGSTYY8 pKa = 10.67DD9 pKa = 2.93ITFQEE14 pKa = 4.47IGSAPRR20 pKa = 11.84DD21 pKa = 3.53LNANWGYY28 pKa = 11.35NEE30 pKa = 4.6LDD32 pKa = 3.62GYY34 pKa = 10.93LYY36 pKa = 9.73TVRR39 pKa = 11.84PGRR42 pKa = 11.84TDD44 pKa = 3.68LYY46 pKa = 10.92RR47 pKa = 11.84VDD49 pKa = 3.6GSGNFVDD56 pKa = 6.75FANLPNTAFDD66 pKa = 3.9GSTAGDD72 pKa = 3.65IFPNGTMVYY81 pKa = 10.37VGDD84 pKa = 4.05ANSWQLIGLATPTAPIDD101 pKa = 4.31LGTLEE106 pKa = 5.35LNQSVNVQDD115 pKa = 3.56IAYY118 pKa = 9.53NPADD122 pKa = 3.52GFLYY126 pKa = 10.45GINRR130 pKa = 11.84NTGRR134 pKa = 11.84AFRR137 pKa = 11.84VDD139 pKa = 3.46GNGGTFGNVTVIEE152 pKa = 4.6FGPAIYY158 pKa = 10.31AGVFASVWFDD168 pKa = 3.15EE169 pKa = 4.55DD170 pKa = 3.46GRR172 pKa = 11.84FYY174 pKa = 10.99GYY176 pKa = 11.39SNTTNNFFLIDD187 pKa = 3.82TTTGNAVPIANGAVDD202 pKa = 4.06EE203 pKa = 5.34GGDD206 pKa = 3.87SDD208 pKa = 4.51GASCRR213 pKa = 11.84GPAPISFGSISGSVYY228 pKa = 11.04DD229 pKa = 5.19DD230 pKa = 4.74LNGSDD235 pKa = 3.45VRR237 pKa = 11.84EE238 pKa = 4.34AGEE241 pKa = 4.0TALGAGIRR249 pKa = 11.84IDD251 pKa = 4.51LYY253 pKa = 11.05YY254 pKa = 11.2DD255 pKa = 3.19NGTPSNYY262 pKa = 9.29TDD264 pKa = 4.73DD265 pKa = 4.9VFAQTTDD272 pKa = 3.3TLSDD276 pKa = 3.38GTYY279 pKa = 9.38TFSDD283 pKa = 3.61LVINQSYY290 pKa = 10.31RR291 pKa = 11.84IQLDD295 pKa = 3.43EE296 pKa = 5.28ADD298 pKa = 4.49PDD300 pKa = 4.0LGPGNLIGTSNPILGVSVAAGAITADD326 pKa = 3.41QDD328 pKa = 3.96FGFDD332 pKa = 3.68PSGSDD337 pKa = 3.94LSLSKK342 pKa = 10.79YY343 pKa = 10.1AAATGTTTPITAVVEE358 pKa = 4.0GDD360 pKa = 3.6TIDD363 pKa = 3.36WVVTITNSGSGSPSGVKK380 pKa = 10.44VIDD383 pKa = 5.25LIPSGFAYY391 pKa = 10.75VSDD394 pKa = 4.63DD395 pKa = 3.77APATGDD401 pKa = 3.71SYY403 pKa = 12.04DD404 pKa = 3.91PSTGLWFVDD413 pKa = 4.9EE414 pKa = 4.53ILSGTSEE421 pKa = 4.38TLTITTTVLGSGDD434 pKa = 3.44FTNYY438 pKa = 10.86AEE440 pKa = 5.07IIYY443 pKa = 10.51SSLPDD448 pKa = 4.5PDD450 pKa = 4.78SDD452 pKa = 4.36PDD454 pKa = 3.68TGRR457 pKa = 11.84LTDD460 pKa = 4.86DD461 pKa = 4.7LSDD464 pKa = 4.17QIGDD468 pKa = 4.05DD469 pKa = 4.45DD470 pKa = 4.05EE471 pKa = 6.8ASYY474 pKa = 11.38AVALQTGSRR483 pKa = 11.84LLSGRR488 pKa = 11.84LFVDD492 pKa = 3.56NGRR495 pKa = 11.84GGGTAHH501 pKa = 7.42DD502 pKa = 4.49GEE504 pKa = 4.79INGAEE509 pKa = 4.23AGSKK513 pKa = 9.77SAQLQILDD521 pKa = 4.01SGGAEE526 pKa = 3.37IGAPQVAADD535 pKa = 4.47GSWSFALSGSYY546 pKa = 10.0SGPLTVRR553 pKa = 11.84ATPLSSHH560 pKa = 5.46ITISEE565 pKa = 3.93ATAGLDD571 pKa = 3.25ALVNADD577 pKa = 3.48PHH579 pKa = 7.89DD580 pKa = 4.27GEE582 pKa = 4.8FTFTPEE588 pKa = 3.97PFGHH592 pKa = 7.01KK593 pKa = 10.01SGLNIGLLEE602 pKa = 4.36TPLLTWNRR610 pKa = 11.84QATVGRR616 pKa = 11.84GQVAEE621 pKa = 4.22LQHH624 pKa = 6.93IYY626 pKa = 9.47TATSQSNVSFTFQNVDD642 pKa = 3.47SAPANAFNASLFLDD656 pKa = 4.15SDD658 pKa = 4.48CDD660 pKa = 3.56GHH662 pKa = 8.09SDD664 pKa = 3.2ITITSPIGVVPEE676 pKa = 3.85QQLCIISRR684 pKa = 11.84VTAASGAGQGSSYY697 pKa = 11.02VYY699 pKa = 10.52QLVAATAFNGTTKK712 pKa = 10.27VHH714 pKa = 5.45TTLNTDD720 pKa = 3.37QVSVTSGTGNVEE732 pKa = 3.65LSKK735 pKa = 9.84TVEE738 pKa = 4.0NQTIGSPEE746 pKa = 3.92GTSNLGRR753 pKa = 11.84AGDD756 pKa = 3.46ILTYY760 pKa = 10.26RR761 pKa = 11.84IYY763 pKa = 11.23LLNTSPQPISNVKK776 pKa = 10.2VYY778 pKa = 11.04DD779 pKa = 3.36RR780 pKa = 11.84TPPYY784 pKa = 9.64TSLSAPIADD793 pKa = 4.03PVTVSPNLTCTVAKK807 pKa = 9.99PVANVTGYY815 pKa = 10.67EE816 pKa = 4.23GDD818 pKa = 4.68LEE820 pKa = 4.65WTCSGSFLPGEE831 pKa = 4.17TGAVEE836 pKa = 4.21FSVAISPP843 pKa = 4.28
MM1 pKa = 7.51TSTGSTYY8 pKa = 10.67DD9 pKa = 2.93ITFQEE14 pKa = 4.47IGSAPRR20 pKa = 11.84DD21 pKa = 3.53LNANWGYY28 pKa = 11.35NEE30 pKa = 4.6LDD32 pKa = 3.62GYY34 pKa = 10.93LYY36 pKa = 9.73TVRR39 pKa = 11.84PGRR42 pKa = 11.84TDD44 pKa = 3.68LYY46 pKa = 10.92RR47 pKa = 11.84VDD49 pKa = 3.6GSGNFVDD56 pKa = 6.75FANLPNTAFDD66 pKa = 3.9GSTAGDD72 pKa = 3.65IFPNGTMVYY81 pKa = 10.37VGDD84 pKa = 4.05ANSWQLIGLATPTAPIDD101 pKa = 4.31LGTLEE106 pKa = 5.35LNQSVNVQDD115 pKa = 3.56IAYY118 pKa = 9.53NPADD122 pKa = 3.52GFLYY126 pKa = 10.45GINRR130 pKa = 11.84NTGRR134 pKa = 11.84AFRR137 pKa = 11.84VDD139 pKa = 3.46GNGGTFGNVTVIEE152 pKa = 4.6FGPAIYY158 pKa = 10.31AGVFASVWFDD168 pKa = 3.15EE169 pKa = 4.55DD170 pKa = 3.46GRR172 pKa = 11.84FYY174 pKa = 10.99GYY176 pKa = 11.39SNTTNNFFLIDD187 pKa = 3.82TTTGNAVPIANGAVDD202 pKa = 4.06EE203 pKa = 5.34GGDD206 pKa = 3.87SDD208 pKa = 4.51GASCRR213 pKa = 11.84GPAPISFGSISGSVYY228 pKa = 11.04DD229 pKa = 5.19DD230 pKa = 4.74LNGSDD235 pKa = 3.45VRR237 pKa = 11.84EE238 pKa = 4.34AGEE241 pKa = 4.0TALGAGIRR249 pKa = 11.84IDD251 pKa = 4.51LYY253 pKa = 11.05YY254 pKa = 11.2DD255 pKa = 3.19NGTPSNYY262 pKa = 9.29TDD264 pKa = 4.73DD265 pKa = 4.9VFAQTTDD272 pKa = 3.3TLSDD276 pKa = 3.38GTYY279 pKa = 9.38TFSDD283 pKa = 3.61LVINQSYY290 pKa = 10.31RR291 pKa = 11.84IQLDD295 pKa = 3.43EE296 pKa = 5.28ADD298 pKa = 4.49PDD300 pKa = 4.0LGPGNLIGTSNPILGVSVAAGAITADD326 pKa = 3.41QDD328 pKa = 3.96FGFDD332 pKa = 3.68PSGSDD337 pKa = 3.94LSLSKK342 pKa = 10.79YY343 pKa = 10.1AAATGTTTPITAVVEE358 pKa = 4.0GDD360 pKa = 3.6TIDD363 pKa = 3.36WVVTITNSGSGSPSGVKK380 pKa = 10.44VIDD383 pKa = 5.25LIPSGFAYY391 pKa = 10.75VSDD394 pKa = 4.63DD395 pKa = 3.77APATGDD401 pKa = 3.71SYY403 pKa = 12.04DD404 pKa = 3.91PSTGLWFVDD413 pKa = 4.9EE414 pKa = 4.53ILSGTSEE421 pKa = 4.38TLTITTTVLGSGDD434 pKa = 3.44FTNYY438 pKa = 10.86AEE440 pKa = 5.07IIYY443 pKa = 10.51SSLPDD448 pKa = 4.5PDD450 pKa = 4.78SDD452 pKa = 4.36PDD454 pKa = 3.68TGRR457 pKa = 11.84LTDD460 pKa = 4.86DD461 pKa = 4.7LSDD464 pKa = 4.17QIGDD468 pKa = 4.05DD469 pKa = 4.45DD470 pKa = 4.05EE471 pKa = 6.8ASYY474 pKa = 11.38AVALQTGSRR483 pKa = 11.84LLSGRR488 pKa = 11.84LFVDD492 pKa = 3.56NGRR495 pKa = 11.84GGGTAHH501 pKa = 7.42DD502 pKa = 4.49GEE504 pKa = 4.79INGAEE509 pKa = 4.23AGSKK513 pKa = 9.77SAQLQILDD521 pKa = 4.01SGGAEE526 pKa = 3.37IGAPQVAADD535 pKa = 4.47GSWSFALSGSYY546 pKa = 10.0SGPLTVRR553 pKa = 11.84ATPLSSHH560 pKa = 5.46ITISEE565 pKa = 3.93ATAGLDD571 pKa = 3.25ALVNADD577 pKa = 3.48PHH579 pKa = 7.89DD580 pKa = 4.27GEE582 pKa = 4.8FTFTPEE588 pKa = 3.97PFGHH592 pKa = 7.01KK593 pKa = 10.01SGLNIGLLEE602 pKa = 4.36TPLLTWNRR610 pKa = 11.84QATVGRR616 pKa = 11.84GQVAEE621 pKa = 4.22LQHH624 pKa = 6.93IYY626 pKa = 9.47TATSQSNVSFTFQNVDD642 pKa = 3.47SAPANAFNASLFLDD656 pKa = 4.15SDD658 pKa = 4.48CDD660 pKa = 3.56GHH662 pKa = 8.09SDD664 pKa = 3.2ITITSPIGVVPEE676 pKa = 3.85QQLCIISRR684 pKa = 11.84VTAASGAGQGSSYY697 pKa = 11.02VYY699 pKa = 10.52QLVAATAFNGTTKK712 pKa = 10.27VHH714 pKa = 5.45TTLNTDD720 pKa = 3.37QVSVTSGTGNVEE732 pKa = 3.65LSKK735 pKa = 9.84TVEE738 pKa = 4.0NQTIGSPEE746 pKa = 3.92GTSNLGRR753 pKa = 11.84AGDD756 pKa = 3.46ILTYY760 pKa = 10.26RR761 pKa = 11.84IYY763 pKa = 11.23LLNTSPQPISNVKK776 pKa = 10.2VYY778 pKa = 11.04DD779 pKa = 3.36RR780 pKa = 11.84TPPYY784 pKa = 9.64TSLSAPIADD793 pKa = 4.03PVTVSPNLTCTVAKK807 pKa = 9.99PVANVTGYY815 pKa = 10.67EE816 pKa = 4.23GDD818 pKa = 4.68LEE820 pKa = 4.65WTCSGSFLPGEE831 pKa = 4.17TGAVEE836 pKa = 4.21FSVAISPP843 pKa = 4.28
Molecular weight: 87.83 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I1TQP8|A0A1I1TQP8_9RHOB L-glutamine ABC transporter membrane protein /L-glutamate ABC transporter membrane protein /L-aspartate ABC transporter membrane protein /L-asparagine ABC transporter membrane protein OS=Sulfitobacter brevis OX=74348 GN=SAMN04488523_101435 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.81IINARR34 pKa = 11.84RR35 pKa = 11.84AQGRR39 pKa = 11.84KK40 pKa = 8.52EE41 pKa = 3.76LSAA44 pKa = 4.93
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.81IINARR34 pKa = 11.84RR35 pKa = 11.84AQGRR39 pKa = 11.84KK40 pKa = 8.52EE41 pKa = 3.76LSAA44 pKa = 4.93
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1253621 |
40 |
2159 |
301.8 |
32.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.139 ± 0.045 | 0.907 ± 0.011 |
6.012 ± 0.031 | 5.666 ± 0.033 |
3.717 ± 0.024 | 8.477 ± 0.042 |
2.081 ± 0.019 | 5.391 ± 0.024 |
3.47 ± 0.031 | 9.918 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.915 ± 0.021 | 2.803 ± 0.021 |
4.903 ± 0.03 | 3.379 ± 0.02 |
6.334 ± 0.038 | 5.363 ± 0.025 |
5.666 ± 0.025 | 7.264 ± 0.033 |
1.368 ± 0.016 | 2.224 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
