
Apis mellifera associated microvirus 13
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.99
Get precalculated fractions of proteins
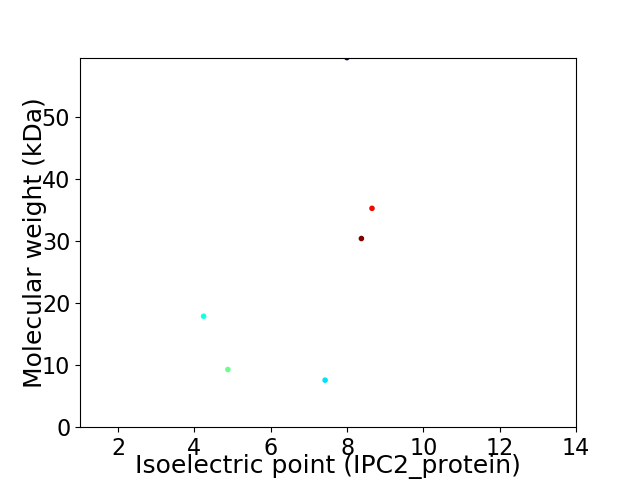
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3Q8U5P9|A0A3Q8U5P9_9VIRU Nonstructural protein OS=Apis mellifera associated microvirus 13 OX=2494740 PE=4 SV=1
MM1 pKa = 7.87AKK3 pKa = 10.34LQMLNAEE10 pKa = 4.2TGEE13 pKa = 4.36IIDD16 pKa = 4.26NNHH19 pKa = 5.89PFIRR23 pKa = 11.84TPYY26 pKa = 10.43NYY28 pKa = 10.45DD29 pKa = 3.01RR30 pKa = 11.84DD31 pKa = 3.93AASLEE36 pKa = 4.23TATVCTEE43 pKa = 3.65EE44 pKa = 4.55TLAQQQFKK52 pKa = 10.78DD53 pKa = 3.44EE54 pKa = 4.42CDD56 pKa = 2.95INTILEE62 pKa = 4.19RR63 pKa = 11.84FGITGEE69 pKa = 4.07LPQNLRR75 pKa = 11.84QPLSADD81 pKa = 3.94FIEE84 pKa = 5.47AYY86 pKa = 10.03DD87 pKa = 3.96YY88 pKa = 9.01KK89 pKa = 10.77TAMDD93 pKa = 5.08SIIEE97 pKa = 4.05AQNAFMEE104 pKa = 4.65LPAKK108 pKa = 10.17IRR110 pKa = 11.84EE111 pKa = 4.24EE112 pKa = 4.17FNNDD116 pKa = 3.1PGQFIQFFEE125 pKa = 4.3QEE127 pKa = 4.2EE128 pKa = 4.04NRR130 pKa = 11.84ARR132 pKa = 11.84AIEE135 pKa = 4.36LGLIQAPRR143 pKa = 11.84EE144 pKa = 4.09PANQAPADD152 pKa = 4.0PTPPTTT158 pKa = 4.07
MM1 pKa = 7.87AKK3 pKa = 10.34LQMLNAEE10 pKa = 4.2TGEE13 pKa = 4.36IIDD16 pKa = 4.26NNHH19 pKa = 5.89PFIRR23 pKa = 11.84TPYY26 pKa = 10.43NYY28 pKa = 10.45DD29 pKa = 3.01RR30 pKa = 11.84DD31 pKa = 3.93AASLEE36 pKa = 4.23TATVCTEE43 pKa = 3.65EE44 pKa = 4.55TLAQQQFKK52 pKa = 10.78DD53 pKa = 3.44EE54 pKa = 4.42CDD56 pKa = 2.95INTILEE62 pKa = 4.19RR63 pKa = 11.84FGITGEE69 pKa = 4.07LPQNLRR75 pKa = 11.84QPLSADD81 pKa = 3.94FIEE84 pKa = 5.47AYY86 pKa = 10.03DD87 pKa = 3.96YY88 pKa = 9.01KK89 pKa = 10.77TAMDD93 pKa = 5.08SIIEE97 pKa = 4.05AQNAFMEE104 pKa = 4.65LPAKK108 pKa = 10.17IRR110 pKa = 11.84EE111 pKa = 4.24EE112 pKa = 4.17FNNDD116 pKa = 3.1PGQFIQFFEE125 pKa = 4.3QEE127 pKa = 4.2EE128 pKa = 4.04NRR130 pKa = 11.84ARR132 pKa = 11.84AIEE135 pKa = 4.36LGLIQAPRR143 pKa = 11.84EE144 pKa = 4.09PANQAPADD152 pKa = 4.0PTPPTTT158 pKa = 4.07
Molecular weight: 17.91 kDa
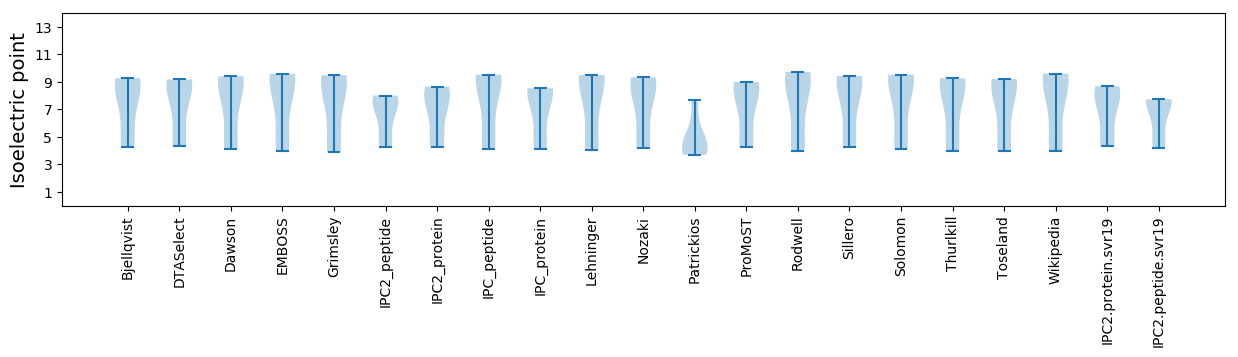
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3S8UTU0|A0A3S8UTU0_9VIRU Replication initiator protein OS=Apis mellifera associated microvirus 13 OX=2494740 PE=4 SV=1
MM1 pKa = 7.26EE2 pKa = 6.08ALMPCYY8 pKa = 10.18HH9 pKa = 7.73PINAYY14 pKa = 7.94KK15 pKa = 8.96TASGEE20 pKa = 4.04IVFTEE25 pKa = 4.05KK26 pKa = 10.61ARR28 pKa = 11.84YY29 pKa = 9.9GDD31 pKa = 3.65TQHH34 pKa = 6.96IKK36 pKa = 10.47LPCGQCIGCRR46 pKa = 11.84LEE48 pKa = 5.65RR49 pKa = 11.84SRR51 pKa = 11.84QWGVRR56 pKa = 11.84CMHH59 pKa = 7.17EE60 pKa = 4.07AQMHH64 pKa = 5.16KK65 pKa = 10.46QNSFVTLTYY74 pKa = 10.37NAEE77 pKa = 4.01NTPQRR82 pKa = 11.84GNLNHH87 pKa = 7.45ADD89 pKa = 3.51FQKK92 pKa = 10.67FMKK95 pKa = 10.4RR96 pKa = 11.84LRR98 pKa = 11.84KK99 pKa = 8.8HH100 pKa = 5.99AKK102 pKa = 8.25TDD104 pKa = 2.53IRR106 pKa = 11.84YY107 pKa = 10.01YY108 pKa = 10.15MGGEE112 pKa = 4.03YY113 pKa = 10.76GPTTWRR119 pKa = 11.84PHH121 pKa = 4.32FHH123 pKa = 6.81ACIFGYY129 pKa = 10.77DD130 pKa = 3.85FADD133 pKa = 3.07KK134 pKa = 10.86LYY136 pKa = 10.54FKK138 pKa = 10.12TGEE141 pKa = 4.2SGEE144 pKa = 4.4KK145 pKa = 9.68IYY147 pKa = 10.5TSKK150 pKa = 9.59TLEE153 pKa = 4.14KK154 pKa = 9.37LWPYY158 pKa = 10.81GFSTIGNVTFQSAAYY173 pKa = 8.08IARR176 pKa = 11.84YY177 pKa = 8.78CVQKK181 pKa = 9.95VTGQAAEE188 pKa = 3.9EE189 pKa = 4.41HH190 pKa = 6.46YY191 pKa = 11.01KK192 pKa = 10.66RR193 pKa = 11.84VDD195 pKa = 3.41DD196 pKa = 3.71QGEE199 pKa = 4.17YY200 pKa = 10.74QLTPEE205 pKa = 4.39YY206 pKa = 11.26NRR208 pKa = 11.84MSLKK212 pKa = 10.3PAIGKK217 pKa = 7.49TWLEE221 pKa = 4.24KK222 pKa = 10.68YY223 pKa = 10.47RR224 pKa = 11.84SDD226 pKa = 4.75VYY228 pKa = 11.53NHH230 pKa = 6.51DD231 pKa = 3.55HH232 pKa = 6.81VIINGVKK239 pKa = 8.56STPPKK244 pKa = 10.88YY245 pKa = 10.09YY246 pKa = 10.67DD247 pKa = 3.69KK248 pKa = 11.19LLKK251 pKa = 10.47RR252 pKa = 11.84WDD254 pKa = 3.59ANRR257 pKa = 11.84LEE259 pKa = 4.1EE260 pKa = 5.04LKK262 pKa = 10.56EE263 pKa = 3.81QRR265 pKa = 11.84EE266 pKa = 4.51ANAQTHH272 pKa = 6.48KK273 pKa = 10.84EE274 pKa = 4.01DD275 pKa = 3.67QTWQRR280 pKa = 11.84LEE282 pKa = 4.05AKK284 pKa = 9.86EE285 pKa = 4.0QFQHH289 pKa = 6.81ARR291 pKa = 11.84IKK293 pKa = 10.79SLLRR297 pKa = 11.84NKK299 pKa = 9.73II300 pKa = 3.43
MM1 pKa = 7.26EE2 pKa = 6.08ALMPCYY8 pKa = 10.18HH9 pKa = 7.73PINAYY14 pKa = 7.94KK15 pKa = 8.96TASGEE20 pKa = 4.04IVFTEE25 pKa = 4.05KK26 pKa = 10.61ARR28 pKa = 11.84YY29 pKa = 9.9GDD31 pKa = 3.65TQHH34 pKa = 6.96IKK36 pKa = 10.47LPCGQCIGCRR46 pKa = 11.84LEE48 pKa = 5.65RR49 pKa = 11.84SRR51 pKa = 11.84QWGVRR56 pKa = 11.84CMHH59 pKa = 7.17EE60 pKa = 4.07AQMHH64 pKa = 5.16KK65 pKa = 10.46QNSFVTLTYY74 pKa = 10.37NAEE77 pKa = 4.01NTPQRR82 pKa = 11.84GNLNHH87 pKa = 7.45ADD89 pKa = 3.51FQKK92 pKa = 10.67FMKK95 pKa = 10.4RR96 pKa = 11.84LRR98 pKa = 11.84KK99 pKa = 8.8HH100 pKa = 5.99AKK102 pKa = 8.25TDD104 pKa = 2.53IRR106 pKa = 11.84YY107 pKa = 10.01YY108 pKa = 10.15MGGEE112 pKa = 4.03YY113 pKa = 10.76GPTTWRR119 pKa = 11.84PHH121 pKa = 4.32FHH123 pKa = 6.81ACIFGYY129 pKa = 10.77DD130 pKa = 3.85FADD133 pKa = 3.07KK134 pKa = 10.86LYY136 pKa = 10.54FKK138 pKa = 10.12TGEE141 pKa = 4.2SGEE144 pKa = 4.4KK145 pKa = 9.68IYY147 pKa = 10.5TSKK150 pKa = 9.59TLEE153 pKa = 4.14KK154 pKa = 9.37LWPYY158 pKa = 10.81GFSTIGNVTFQSAAYY173 pKa = 8.08IARR176 pKa = 11.84YY177 pKa = 8.78CVQKK181 pKa = 9.95VTGQAAEE188 pKa = 3.9EE189 pKa = 4.41HH190 pKa = 6.46YY191 pKa = 11.01KK192 pKa = 10.66RR193 pKa = 11.84VDD195 pKa = 3.41DD196 pKa = 3.71QGEE199 pKa = 4.17YY200 pKa = 10.74QLTPEE205 pKa = 4.39YY206 pKa = 11.26NRR208 pKa = 11.84MSLKK212 pKa = 10.3PAIGKK217 pKa = 7.49TWLEE221 pKa = 4.24KK222 pKa = 10.68YY223 pKa = 10.47RR224 pKa = 11.84SDD226 pKa = 4.75VYY228 pKa = 11.53NHH230 pKa = 6.51DD231 pKa = 3.55HH232 pKa = 6.81VIINGVKK239 pKa = 8.56STPPKK244 pKa = 10.88YY245 pKa = 10.09YY246 pKa = 10.67DD247 pKa = 3.69KK248 pKa = 11.19LLKK251 pKa = 10.47RR252 pKa = 11.84WDD254 pKa = 3.59ANRR257 pKa = 11.84LEE259 pKa = 4.1EE260 pKa = 5.04LKK262 pKa = 10.56EE263 pKa = 3.81QRR265 pKa = 11.84EE266 pKa = 4.51ANAQTHH272 pKa = 6.48KK273 pKa = 10.84EE274 pKa = 4.01DD275 pKa = 3.67QTWQRR280 pKa = 11.84LEE282 pKa = 4.05AKK284 pKa = 9.86EE285 pKa = 4.0QFQHH289 pKa = 6.81ARR291 pKa = 11.84IKK293 pKa = 10.79SLLRR297 pKa = 11.84NKK299 pKa = 9.73II300 pKa = 3.43
Molecular weight: 35.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1428 |
67 |
539 |
238.0 |
26.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.294 ± 1.383 | 0.77 ± 0.464 |
4.692 ± 0.607 | 5.882 ± 1.352 |
3.992 ± 0.938 | 6.162 ± 0.652 |
2.171 ± 0.801 | 5.882 ± 0.609 |
4.902 ± 1.404 | 7.143 ± 0.527 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.591 ± 0.167 | 5.602 ± 0.555 |
5.112 ± 1.069 | 6.793 ± 0.839 |
5.602 ± 0.363 | 4.972 ± 0.937 |
8.333 ± 0.62 | 3.852 ± 0.682 |
1.331 ± 0.382 | 3.922 ± 0.944 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
