
Clostridium sp. D5
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Clostridiaceae; Clostridium; unclassified Clostridium
Average proteome isoelectric point is 6.02
Get precalculated fractions of proteins
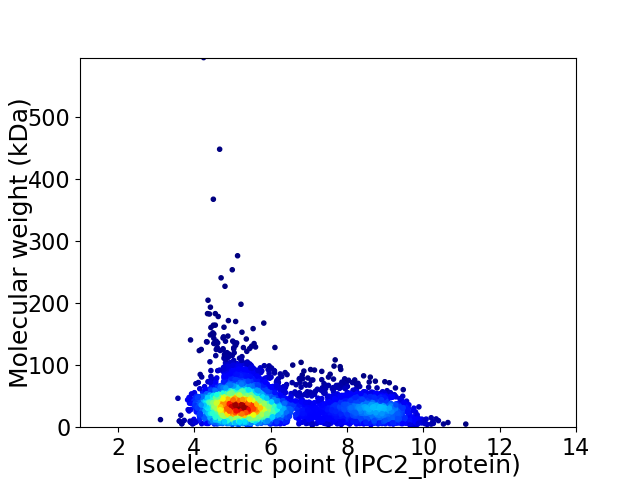
Virtual 2D-PAGE plot for 4496 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F0YVL7|F0YVL7_9CLOT Transposase IS66 (Fragment) OS=Clostridium sp. D5 OX=556261 GN=HMPREF0240_01188 PE=4 SV=1
MM1 pKa = 7.15IAGMAAGCGSGSEE14 pKa = 4.4SKK16 pKa = 10.82KK17 pKa = 10.5SDD19 pKa = 3.06SSEE22 pKa = 4.04DD23 pKa = 3.34GKK25 pKa = 11.26EE26 pKa = 4.63VVLTWQSYY34 pKa = 10.57DD35 pKa = 3.92SYY37 pKa = 11.87DD38 pKa = 3.46KK39 pKa = 11.37YY40 pKa = 11.39EE41 pKa = 4.1KK42 pKa = 10.98VVDD45 pKa = 3.86AFEE48 pKa = 5.18KK49 pKa = 10.45EE50 pKa = 4.02NPDD53 pKa = 2.86IKK55 pKa = 10.67IEE57 pKa = 4.05FEE59 pKa = 4.15EE60 pKa = 4.71VSDD63 pKa = 4.23FSTKK67 pKa = 10.05ILTEE71 pKa = 3.92ATAGDD76 pKa = 4.44LPDD79 pKa = 6.27LINCNTGTTQLLAEE93 pKa = 4.71SGALQKK99 pKa = 10.66FDD101 pKa = 3.97TEE103 pKa = 3.95EE104 pKa = 4.8LEE106 pKa = 4.51ADD108 pKa = 3.69EE109 pKa = 5.36EE110 pKa = 5.0YY111 pKa = 11.13KK112 pKa = 10.66FDD114 pKa = 5.4DD115 pKa = 3.79FWDD118 pKa = 3.9AAKK121 pKa = 9.76TYY123 pKa = 8.48CTYY126 pKa = 11.29DD127 pKa = 3.81DD128 pKa = 3.47EE129 pKa = 5.19WYY131 pKa = 10.58ALPLDD136 pKa = 4.04GGNYY140 pKa = 7.97GWVYY144 pKa = 10.07NVDD147 pKa = 3.48MFRR150 pKa = 11.84EE151 pKa = 4.29CGIEE155 pKa = 3.99VPEE158 pKa = 5.29EE159 pKa = 3.96GFTWKK164 pKa = 10.25EE165 pKa = 3.65FEE167 pKa = 4.41EE168 pKa = 4.71ACSLLNEE175 pKa = 4.59NKK177 pKa = 10.17DD178 pKa = 3.54KK179 pKa = 11.53LGIEE183 pKa = 4.42YY184 pKa = 7.99PTIFNDD190 pKa = 3.66LSSSIDD196 pKa = 3.23MMYY199 pKa = 10.86PLITEE204 pKa = 4.56AGGEE208 pKa = 4.09YY209 pKa = 10.69LNADD213 pKa = 4.14GTCAWNSEE221 pKa = 4.18AVVTAFEE228 pKa = 4.57WVQSLVAEE236 pKa = 5.22GYY238 pKa = 9.72IPSIEE243 pKa = 4.13KK244 pKa = 10.83LGDD247 pKa = 3.73GYY249 pKa = 11.38DD250 pKa = 3.53ALITKK255 pKa = 9.11FNAGQIAMCRR265 pKa = 11.84VALWNSTYY273 pKa = 11.16LEE275 pKa = 4.3DD276 pKa = 3.74TVNWKK281 pKa = 9.59VMNAPRR287 pKa = 11.84GNDD290 pKa = 4.19GIQSEE295 pKa = 4.84VLFLNGIGISNTCEE309 pKa = 3.84NYY311 pKa = 10.12DD312 pKa = 3.49AALKK316 pKa = 9.23FVKK319 pKa = 10.46YY320 pKa = 8.73LTSEE324 pKa = 4.14EE325 pKa = 4.07GLAMYY330 pKa = 10.54LKK332 pKa = 10.8DD333 pKa = 3.49NTSPQIAVRR342 pKa = 11.84RR343 pKa = 11.84SQADD347 pKa = 3.25LSVAMFDD354 pKa = 4.5EE355 pKa = 4.3KK356 pKa = 11.18HH357 pKa = 7.22DD358 pKa = 3.75MQLYY362 pKa = 7.72NTGLEE367 pKa = 4.06YY368 pKa = 10.68AGYY371 pKa = 9.99IDD373 pKa = 4.84LTNTFADD380 pKa = 3.4QQTIIGQSFDD390 pKa = 4.75EE391 pKa = 4.5IWHH394 pKa = 6.46NDD396 pKa = 2.67ADD398 pKa = 3.9IQKK401 pKa = 8.15TLDD404 pKa = 3.64SLVEE408 pKa = 3.98QLDD411 pKa = 3.86GLLTQQ416 pKa = 4.71
MM1 pKa = 7.15IAGMAAGCGSGSEE14 pKa = 4.4SKK16 pKa = 10.82KK17 pKa = 10.5SDD19 pKa = 3.06SSEE22 pKa = 4.04DD23 pKa = 3.34GKK25 pKa = 11.26EE26 pKa = 4.63VVLTWQSYY34 pKa = 10.57DD35 pKa = 3.92SYY37 pKa = 11.87DD38 pKa = 3.46KK39 pKa = 11.37YY40 pKa = 11.39EE41 pKa = 4.1KK42 pKa = 10.98VVDD45 pKa = 3.86AFEE48 pKa = 5.18KK49 pKa = 10.45EE50 pKa = 4.02NPDD53 pKa = 2.86IKK55 pKa = 10.67IEE57 pKa = 4.05FEE59 pKa = 4.15EE60 pKa = 4.71VSDD63 pKa = 4.23FSTKK67 pKa = 10.05ILTEE71 pKa = 3.92ATAGDD76 pKa = 4.44LPDD79 pKa = 6.27LINCNTGTTQLLAEE93 pKa = 4.71SGALQKK99 pKa = 10.66FDD101 pKa = 3.97TEE103 pKa = 3.95EE104 pKa = 4.8LEE106 pKa = 4.51ADD108 pKa = 3.69EE109 pKa = 5.36EE110 pKa = 5.0YY111 pKa = 11.13KK112 pKa = 10.66FDD114 pKa = 5.4DD115 pKa = 3.79FWDD118 pKa = 3.9AAKK121 pKa = 9.76TYY123 pKa = 8.48CTYY126 pKa = 11.29DD127 pKa = 3.81DD128 pKa = 3.47EE129 pKa = 5.19WYY131 pKa = 10.58ALPLDD136 pKa = 4.04GGNYY140 pKa = 7.97GWVYY144 pKa = 10.07NVDD147 pKa = 3.48MFRR150 pKa = 11.84EE151 pKa = 4.29CGIEE155 pKa = 3.99VPEE158 pKa = 5.29EE159 pKa = 3.96GFTWKK164 pKa = 10.25EE165 pKa = 3.65FEE167 pKa = 4.41EE168 pKa = 4.71ACSLLNEE175 pKa = 4.59NKK177 pKa = 10.17DD178 pKa = 3.54KK179 pKa = 11.53LGIEE183 pKa = 4.42YY184 pKa = 7.99PTIFNDD190 pKa = 3.66LSSSIDD196 pKa = 3.23MMYY199 pKa = 10.86PLITEE204 pKa = 4.56AGGEE208 pKa = 4.09YY209 pKa = 10.69LNADD213 pKa = 4.14GTCAWNSEE221 pKa = 4.18AVVTAFEE228 pKa = 4.57WVQSLVAEE236 pKa = 5.22GYY238 pKa = 9.72IPSIEE243 pKa = 4.13KK244 pKa = 10.83LGDD247 pKa = 3.73GYY249 pKa = 11.38DD250 pKa = 3.53ALITKK255 pKa = 9.11FNAGQIAMCRR265 pKa = 11.84VALWNSTYY273 pKa = 11.16LEE275 pKa = 4.3DD276 pKa = 3.74TVNWKK281 pKa = 9.59VMNAPRR287 pKa = 11.84GNDD290 pKa = 4.19GIQSEE295 pKa = 4.84VLFLNGIGISNTCEE309 pKa = 3.84NYY311 pKa = 10.12DD312 pKa = 3.49AALKK316 pKa = 9.23FVKK319 pKa = 10.46YY320 pKa = 8.73LTSEE324 pKa = 4.14EE325 pKa = 4.07GLAMYY330 pKa = 10.54LKK332 pKa = 10.8DD333 pKa = 3.49NTSPQIAVRR342 pKa = 11.84RR343 pKa = 11.84SQADD347 pKa = 3.25LSVAMFDD354 pKa = 4.5EE355 pKa = 4.3KK356 pKa = 11.18HH357 pKa = 7.22DD358 pKa = 3.75MQLYY362 pKa = 7.72NTGLEE367 pKa = 4.06YY368 pKa = 10.68AGYY371 pKa = 9.99IDD373 pKa = 4.84LTNTFADD380 pKa = 3.4QQTIIGQSFDD390 pKa = 4.75EE391 pKa = 4.5IWHH394 pKa = 6.46NDD396 pKa = 2.67ADD398 pKa = 3.9IQKK401 pKa = 8.15TLDD404 pKa = 3.64SLVEE408 pKa = 3.98QLDD411 pKa = 3.86GLLTQQ416 pKa = 4.71
Molecular weight: 46.59 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F0YT62|F0YT62_9CLOT Alpha-L-fucosidase OS=Clostridium sp. D5 OX=556261 GN=HMPREF0240_00319 PE=4 SV=1
MM1 pKa = 7.67KK2 pKa = 8.71MTFQPKK8 pKa = 7.43TRR10 pKa = 11.84QRR12 pKa = 11.84AKK14 pKa = 9.26VHH16 pKa = 5.59GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTPGGRR28 pKa = 11.84KK29 pKa = 8.8VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.18GRR39 pKa = 11.84KK40 pKa = 8.18QLSAA44 pKa = 3.9
MM1 pKa = 7.67KK2 pKa = 8.71MTFQPKK8 pKa = 7.43TRR10 pKa = 11.84QRR12 pKa = 11.84AKK14 pKa = 9.26VHH16 pKa = 5.59GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTPGGRR28 pKa = 11.84KK29 pKa = 8.8VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.18GRR39 pKa = 11.84KK40 pKa = 8.18QLSAA44 pKa = 3.9
Molecular weight: 5.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1527431 |
29 |
5529 |
339.7 |
38.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.604 ± 0.037 | 1.549 ± 0.015 |
5.474 ± 0.029 | 7.683 ± 0.043 |
4.098 ± 0.028 | 7.452 ± 0.033 |
1.728 ± 0.017 | 7.361 ± 0.035 |
6.453 ± 0.026 | 9.033 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.277 ± 0.021 | 4.118 ± 0.022 |
3.436 ± 0.021 | 3.279 ± 0.021 |
4.416 ± 0.03 | 5.757 ± 0.03 |
5.293 ± 0.029 | 6.881 ± 0.034 |
0.998 ± 0.013 | 4.11 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
