
Bos taurus papillomavirus 23
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Xipapillomavirus; Xipapillomavirus 1
Average proteome isoelectric point is 6.01
Get precalculated fractions of proteins
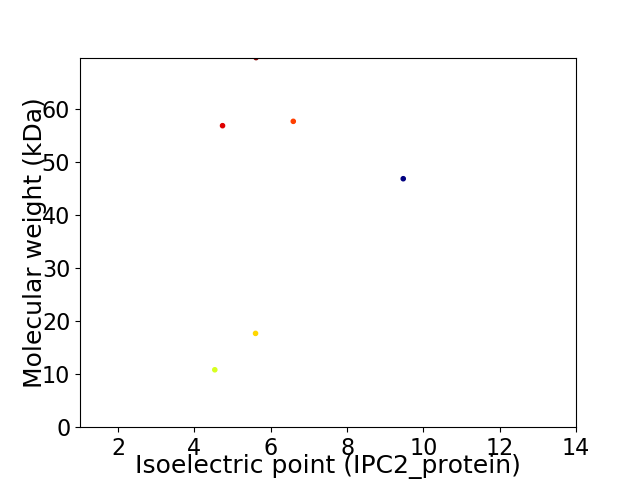
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L6KTE6|A0A1L6KTE6_BPV3 Protein E7 OS=Bos taurus papillomavirus 23 OX=2758958 GN=E7 PE=3 SV=1
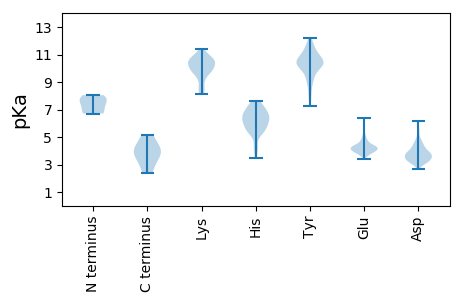
MM1 pKa = 7.65KK2 pKa = 10.33GQDD5 pKa = 3.22VTLQGIAVEE14 pKa = 4.27LKK16 pKa = 10.45EE17 pKa = 4.22VVSPINLACDD27 pKa = 3.47EE28 pKa = 4.64EE29 pKa = 5.44IEE31 pKa = 4.31TEE33 pKa = 4.36EE34 pKa = 4.12VDD36 pKa = 4.41GPAPYY41 pKa = 10.23AVEE44 pKa = 4.01AVCYY48 pKa = 10.5VCEE51 pKa = 4.0QPLRR55 pKa = 11.84LALVADD61 pKa = 4.3SEE63 pKa = 5.14GILQFHH69 pKa = 6.92HH70 pKa = 7.21LLLEE74 pKa = 4.18NLNLLCASCSRR85 pKa = 11.84EE86 pKa = 4.02VFCNRR91 pKa = 11.84RR92 pKa = 11.84PQRR95 pKa = 11.84NGSS98 pKa = 3.34
MM1 pKa = 7.65KK2 pKa = 10.33GQDD5 pKa = 3.22VTLQGIAVEE14 pKa = 4.27LKK16 pKa = 10.45EE17 pKa = 4.22VVSPINLACDD27 pKa = 3.47EE28 pKa = 4.64EE29 pKa = 5.44IEE31 pKa = 4.31TEE33 pKa = 4.36EE34 pKa = 4.12VDD36 pKa = 4.41GPAPYY41 pKa = 10.23AVEE44 pKa = 4.01AVCYY48 pKa = 10.5VCEE51 pKa = 4.0QPLRR55 pKa = 11.84LALVADD61 pKa = 4.3SEE63 pKa = 5.14GILQFHH69 pKa = 6.92HH70 pKa = 7.21LLLEE74 pKa = 4.18NLNLLCASCSRR85 pKa = 11.84EE86 pKa = 4.02VFCNRR91 pKa = 11.84RR92 pKa = 11.84PQRR95 pKa = 11.84NGSS98 pKa = 3.34
Molecular weight: 10.81 kDa
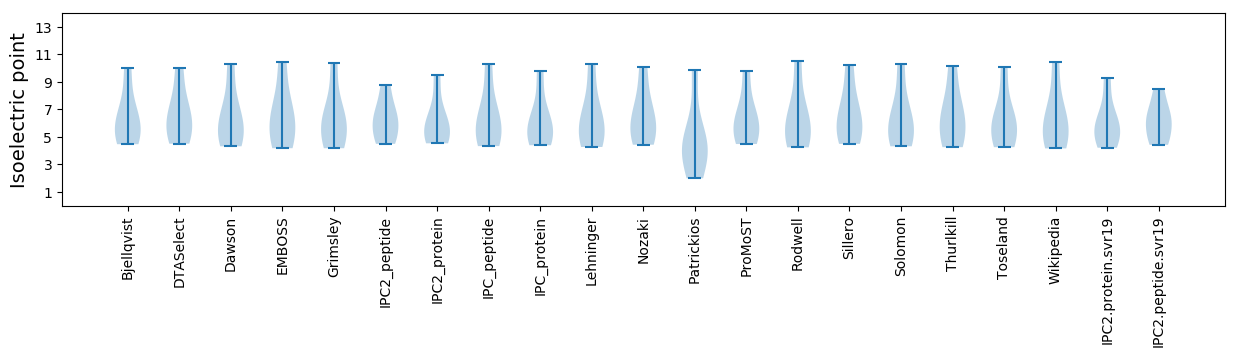
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L6KTE3|A0A1L6KTE3_BPV3 Major capsid protein L1 OS=Bos taurus papillomavirus 23 OX=2758958 GN=L1 PE=3 SV=1
MM1 pKa = 7.79EE2 pKa = 4.97NLQARR7 pKa = 11.84FDD9 pKa = 3.93AVQGQLLEE17 pKa = 4.75IYY19 pKa = 10.15EE20 pKa = 4.16QDD22 pKa = 3.81TNDD25 pKa = 3.62LAVQVTYY32 pKa = 7.74WTLVRR37 pKa = 11.84QEE39 pKa = 3.8QALYY43 pKa = 10.66YY44 pKa = 9.43YY45 pKa = 10.77ARR47 pKa = 11.84QQGITRR53 pKa = 11.84LGLYY57 pKa = 7.98QVPPTRR63 pKa = 11.84VSEE66 pKa = 4.42KK67 pKa = 9.86KK68 pKa = 10.65AKK70 pKa = 10.2DD71 pKa = 3.76AIKK74 pKa = 8.32MTLFLQSLQKK84 pKa = 10.63SEE86 pKa = 4.03FAKK89 pKa = 10.79LSWSLTEE96 pKa = 3.91TSLEE100 pKa = 4.08RR101 pKa = 11.84FLAAPEE107 pKa = 3.93NTFKK111 pKa = 11.01KK112 pKa = 9.98GGQHH116 pKa = 4.2VTVIYY121 pKa = 10.14DD122 pKa = 3.7ANAYY126 pKa = 10.24NAMEE130 pKa = 3.96YY131 pKa = 9.3TLWGEE136 pKa = 3.91IFHH139 pKa = 7.37VDD141 pKa = 4.84EE142 pKa = 4.42TDD144 pKa = 3.05TWHH147 pKa = 7.23KK148 pKa = 10.76SYY150 pKa = 11.63SDD152 pKa = 3.24VDD154 pKa = 3.59YY155 pKa = 11.59DD156 pKa = 4.01GIYY159 pKa = 8.93YY160 pKa = 10.06TDD162 pKa = 3.33VQGNKK167 pKa = 8.55VYY169 pKa = 10.79YY170 pKa = 11.06VNFSDD175 pKa = 5.27DD176 pKa = 3.35AKK178 pKa = 10.63LYY180 pKa = 11.06SKK182 pKa = 10.19MGLWEE187 pKa = 3.75VRR189 pKa = 11.84YY190 pKa = 10.0QNTVLSPPVTSSVPAGSPRR209 pKa = 11.84GRR211 pKa = 11.84HH212 pKa = 5.19GPQTGSSTSGHH223 pKa = 5.65KK224 pKa = 9.39TSLHH228 pKa = 5.47TRR230 pKa = 11.84SQSSEE235 pKa = 3.65SDD237 pKa = 3.32SRR239 pKa = 11.84DD240 pKa = 2.92SRR242 pKa = 11.84GRR244 pKa = 11.84SRR246 pKa = 11.84SSSSSRR252 pKa = 11.84TRR254 pKa = 11.84SRR256 pKa = 11.84SRR258 pKa = 11.84SRR260 pKa = 11.84SRR262 pKa = 11.84SPSGSHH268 pKa = 4.24TRR270 pKa = 11.84EE271 pKa = 3.72RR272 pKa = 11.84TPQLQTGRR280 pKa = 11.84PPGGGRR286 pKa = 11.84GGRR289 pKa = 11.84RR290 pKa = 11.84TGQQRR295 pKa = 11.84DD296 pKa = 3.42RR297 pKa = 11.84RR298 pKa = 11.84RR299 pKa = 11.84PSPPSPGAVGTRR311 pKa = 11.84LRR313 pKa = 11.84TPEE316 pKa = 3.78RR317 pKa = 11.84KK318 pKa = 9.72AKK320 pKa = 9.94GRR322 pKa = 11.84LVQLIEE328 pKa = 4.2EE329 pKa = 4.49ALDD332 pKa = 3.83PPVLLLQGGANTLKK346 pKa = 10.43SFRR349 pKa = 11.84RR350 pKa = 11.84RR351 pKa = 11.84TSKK354 pKa = 10.5SYY356 pKa = 9.4PHH358 pKa = 7.36KK359 pKa = 8.32FTCMSTCWTWASKK372 pKa = 9.22TCTVKK377 pKa = 10.45SGHH380 pKa = 6.37RR381 pKa = 11.84MLIAFQNSEE390 pKa = 3.69QRR392 pKa = 11.84TSFLSTVRR400 pKa = 11.84LPKK403 pKa = 10.63GVTCVKK409 pKa = 10.57GAFDD413 pKa = 3.92SLL415 pKa = 3.89
MM1 pKa = 7.79EE2 pKa = 4.97NLQARR7 pKa = 11.84FDD9 pKa = 3.93AVQGQLLEE17 pKa = 4.75IYY19 pKa = 10.15EE20 pKa = 4.16QDD22 pKa = 3.81TNDD25 pKa = 3.62LAVQVTYY32 pKa = 7.74WTLVRR37 pKa = 11.84QEE39 pKa = 3.8QALYY43 pKa = 10.66YY44 pKa = 9.43YY45 pKa = 10.77ARR47 pKa = 11.84QQGITRR53 pKa = 11.84LGLYY57 pKa = 7.98QVPPTRR63 pKa = 11.84VSEE66 pKa = 4.42KK67 pKa = 9.86KK68 pKa = 10.65AKK70 pKa = 10.2DD71 pKa = 3.76AIKK74 pKa = 8.32MTLFLQSLQKK84 pKa = 10.63SEE86 pKa = 4.03FAKK89 pKa = 10.79LSWSLTEE96 pKa = 3.91TSLEE100 pKa = 4.08RR101 pKa = 11.84FLAAPEE107 pKa = 3.93NTFKK111 pKa = 11.01KK112 pKa = 9.98GGQHH116 pKa = 4.2VTVIYY121 pKa = 10.14DD122 pKa = 3.7ANAYY126 pKa = 10.24NAMEE130 pKa = 3.96YY131 pKa = 9.3TLWGEE136 pKa = 3.91IFHH139 pKa = 7.37VDD141 pKa = 4.84EE142 pKa = 4.42TDD144 pKa = 3.05TWHH147 pKa = 7.23KK148 pKa = 10.76SYY150 pKa = 11.63SDD152 pKa = 3.24VDD154 pKa = 3.59YY155 pKa = 11.59DD156 pKa = 4.01GIYY159 pKa = 8.93YY160 pKa = 10.06TDD162 pKa = 3.33VQGNKK167 pKa = 8.55VYY169 pKa = 10.79YY170 pKa = 11.06VNFSDD175 pKa = 5.27DD176 pKa = 3.35AKK178 pKa = 10.63LYY180 pKa = 11.06SKK182 pKa = 10.19MGLWEE187 pKa = 3.75VRR189 pKa = 11.84YY190 pKa = 10.0QNTVLSPPVTSSVPAGSPRR209 pKa = 11.84GRR211 pKa = 11.84HH212 pKa = 5.19GPQTGSSTSGHH223 pKa = 5.65KK224 pKa = 9.39TSLHH228 pKa = 5.47TRR230 pKa = 11.84SQSSEE235 pKa = 3.65SDD237 pKa = 3.32SRR239 pKa = 11.84DD240 pKa = 2.92SRR242 pKa = 11.84GRR244 pKa = 11.84SRR246 pKa = 11.84SSSSSRR252 pKa = 11.84TRR254 pKa = 11.84SRR256 pKa = 11.84SRR258 pKa = 11.84SRR260 pKa = 11.84SRR262 pKa = 11.84SPSGSHH268 pKa = 4.24TRR270 pKa = 11.84EE271 pKa = 3.72RR272 pKa = 11.84TPQLQTGRR280 pKa = 11.84PPGGGRR286 pKa = 11.84GGRR289 pKa = 11.84RR290 pKa = 11.84TGQQRR295 pKa = 11.84DD296 pKa = 3.42RR297 pKa = 11.84RR298 pKa = 11.84RR299 pKa = 11.84PSPPSPGAVGTRR311 pKa = 11.84LRR313 pKa = 11.84TPEE316 pKa = 3.78RR317 pKa = 11.84KK318 pKa = 9.72AKK320 pKa = 9.94GRR322 pKa = 11.84LVQLIEE328 pKa = 4.2EE329 pKa = 4.49ALDD332 pKa = 3.83PPVLLLQGGANTLKK346 pKa = 10.43SFRR349 pKa = 11.84RR350 pKa = 11.84RR351 pKa = 11.84TSKK354 pKa = 10.5SYY356 pKa = 9.4PHH358 pKa = 7.36KK359 pKa = 8.32FTCMSTCWTWASKK372 pKa = 9.22TCTVKK377 pKa = 10.45SGHH380 pKa = 6.37RR381 pKa = 11.84MLIAFQNSEE390 pKa = 3.69QRR392 pKa = 11.84TSFLSTVRR400 pKa = 11.84LPKK403 pKa = 10.63GVTCVKK409 pKa = 10.57GAFDD413 pKa = 3.92SLL415 pKa = 3.89
Molecular weight: 46.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2311 |
98 |
612 |
385.2 |
43.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.798 ± 0.641 | 1.774 ± 0.503 |
6.015 ± 0.476 | 6.447 ± 0.647 |
4.543 ± 0.501 | 7.183 ± 0.77 |
2.207 ± 0.124 | 4.587 ± 0.707 |
4.89 ± 0.571 | 8.481 ± 0.728 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.688 ± 0.245 | 4.587 ± 0.665 |
6.664 ± 1.123 | 4.587 ± 0.534 |
6.577 ± 0.932 | 8.005 ± 1.095 |
6.188 ± 0.782 | 5.452 ± 0.766 |
1.341 ± 0.201 | 2.986 ± 0.359 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
