
Halodesulfovibrio spirochaetisodalis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfovibrionales; Desulfovibrionaceae; Halodesulfovibrio
Average proteome isoelectric point is 6.18
Get precalculated fractions of proteins
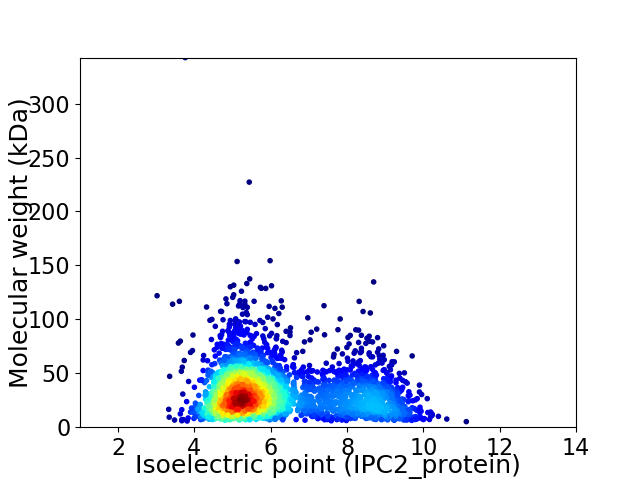
Virtual 2D-PAGE plot for 2803 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B7XAI0|A0A1B7XAI0_9DELT Uncharacterized protein OS=Halodesulfovibrio spirochaetisodalis OX=1560234 GN=SP90_12790 PE=4 SV=1
GG1 pKa = 7.65EE2 pKa = 5.96DD3 pKa = 3.66GDD5 pKa = 4.42DD6 pKa = 4.12TITTTGSAYY15 pKa = 9.46QHH17 pKa = 5.19VQGGNGNDD25 pKa = 4.04TIRR28 pKa = 11.84TGSANDD34 pKa = 4.48AIIGGADD41 pKa = 4.09NDD43 pKa = 4.82TIWAGEE49 pKa = 4.09GDD51 pKa = 3.52NSIWGGTGEE60 pKa = 4.96DD61 pKa = 4.48EE62 pKa = 4.19IHH64 pKa = 6.6SGSGNDD70 pKa = 4.14TIDD73 pKa = 4.35AGDD76 pKa = 4.47DD77 pKa = 3.68NDD79 pKa = 5.1TIWAGDD85 pKa = 3.56GTNTIDD91 pKa = 3.81AGKK94 pKa = 10.76GNDD97 pKa = 4.1IIHH100 pKa = 7.0AGANADD106 pKa = 3.57TYY108 pKa = 11.02KK109 pKa = 10.58YY110 pKa = 10.95SKK112 pKa = 11.21GDD114 pKa = 3.46GHH116 pKa = 7.13DD117 pKa = 3.46VIYY120 pKa = 10.73DD121 pKa = 3.74NEE123 pKa = 4.57SGAVTADD130 pKa = 3.21HH131 pKa = 6.53TLDD134 pKa = 4.58AIRR137 pKa = 11.84FSSDD141 pKa = 2.07IAYY144 pKa = 10.51NDD146 pKa = 3.5LQFAIDD152 pKa = 4.68GNDD155 pKa = 3.6LLITFKK161 pKa = 11.0NSPEE165 pKa = 3.59DD166 pKa = 3.45SIRR169 pKa = 11.84IKK171 pKa = 10.81DD172 pKa = 3.28AATNPEE178 pKa = 4.0HH179 pKa = 6.91AVEE182 pKa = 5.47KK183 pKa = 10.6ILIEE187 pKa = 3.94NRR189 pKa = 11.84YY190 pKa = 8.97NASQVSTYY198 pKa = 9.95YY199 pKa = 10.69VEE201 pKa = 4.15NAAIRR206 pKa = 11.84HH207 pKa = 4.49TTYY210 pKa = 11.29SDD212 pKa = 3.51GDD214 pKa = 3.77DD215 pKa = 4.09TISVPVSSKK224 pKa = 8.1ITKK227 pKa = 10.13ISAGDD232 pKa = 3.66GNDD235 pKa = 3.18TVTSGDD241 pKa = 3.34SSVIIDD247 pKa = 4.1GGAGNDD253 pKa = 4.0TITTGSKK260 pKa = 10.43SDD262 pKa = 3.86VISGGTGNDD271 pKa = 3.73TINAGGGSDD280 pKa = 3.56TLTGGSGSDD289 pKa = 3.06TYY291 pKa = 11.11IFNIGDD297 pKa = 3.49GHH299 pKa = 5.95DD300 pKa = 4.03TIIEE304 pKa = 4.03ASVDD308 pKa = 3.81PTCSCVDD315 pKa = 4.07TISFGAGITSSDD327 pKa = 3.43IIYY330 pKa = 10.55KK331 pKa = 10.12VDD333 pKa = 3.53GNDD336 pKa = 4.95LIICIKK342 pKa = 10.55GHH344 pKa = 6.98DD345 pKa = 4.08GDD347 pKa = 4.67SIRR350 pKa = 11.84ITGALNGASHH360 pKa = 7.45GIEE363 pKa = 4.16TITFASGTTITLNTVISCHH382 pKa = 6.29NYY384 pKa = 8.89TDD386 pKa = 3.74ASDD389 pKa = 4.21TIVVPADD396 pKa = 3.36KK397 pKa = 10.91YY398 pKa = 10.64VVVINAGGGNDD409 pKa = 4.27TITGGDD415 pKa = 3.31TVSIINGGTGNDD427 pKa = 4.02SISTGGKK434 pKa = 10.02DD435 pKa = 3.28DD436 pKa = 3.92TLTGGEE442 pKa = 4.35GDD444 pKa = 3.72DD445 pKa = 4.49TINAGGGNDD454 pKa = 4.25TITGGTGSDD463 pKa = 3.41TLTGGSGSDD472 pKa = 3.06TYY474 pKa = 11.11IFNIGDD480 pKa = 3.49GHH482 pKa = 5.95DD483 pKa = 4.03TIIEE487 pKa = 4.03ASVDD491 pKa = 3.74PTCTCVDD498 pKa = 4.4TISFGAGITSSDD510 pKa = 3.43IIYY513 pKa = 10.55KK514 pKa = 10.12VDD516 pKa = 3.53GNDD519 pKa = 4.95LIICIKK525 pKa = 10.55GHH527 pKa = 6.98DD528 pKa = 4.08GDD530 pKa = 4.67SIRR533 pKa = 11.84ITGALNGASHH543 pKa = 7.45GIEE546 pKa = 4.16TITFASGTSITLNTVISCHH565 pKa = 6.29NYY567 pKa = 8.89TDD569 pKa = 3.74ASDD572 pKa = 4.21TIVVPADD579 pKa = 3.36KK580 pKa = 10.91YY581 pKa = 10.64VVVINAGGGNDD592 pKa = 4.27TITGGDD598 pKa = 3.31TVSIINGGTGNDD610 pKa = 4.02SISTGGKK617 pKa = 10.04DD618 pKa = 4.33DD619 pKa = 3.92ILTGGEE625 pKa = 4.02GDD627 pKa = 3.7DD628 pKa = 4.49TINAGGGNDD637 pKa = 4.13IITGGTGSDD646 pKa = 3.41TLTGGSGSDD655 pKa = 3.06TYY657 pKa = 11.11IFNIGDD663 pKa = 3.49GHH665 pKa = 5.95DD666 pKa = 4.03TIIEE670 pKa = 4.03ASVDD674 pKa = 3.81PTCSCVDD681 pKa = 4.07TISFGAGITSSDD693 pKa = 3.43IIYY696 pKa = 10.55KK697 pKa = 10.12VDD699 pKa = 3.53GNDD702 pKa = 4.95LIICIKK708 pKa = 10.55GHH710 pKa = 6.98DD711 pKa = 4.08GDD713 pKa = 4.67SIRR716 pKa = 11.84ITGALNGTSSFGVEE730 pKa = 4.15NISFTDD736 pKa = 3.87GTSIALTTVISCQNYY751 pKa = 8.51TDD753 pKa = 3.92ASDD756 pKa = 4.2TIVVPADD763 pKa = 3.34KK764 pKa = 10.77YY765 pKa = 10.03VAVINAGGGNDD776 pKa = 4.27TITGGDD782 pKa = 3.28TASIINGGAGNDD794 pKa = 4.16SISTGAKK801 pKa = 10.11DD802 pKa = 4.98DD803 pKa = 3.95ILNGGIGNDD812 pKa = 3.95TINAGGGNDD821 pKa = 4.25TITGGTGTDD830 pKa = 3.48TLIGGSGSDD839 pKa = 3.12TYY841 pKa = 11.11IFNIGDD847 pKa = 3.49GHH849 pKa = 5.95DD850 pKa = 4.03TIIEE854 pKa = 4.03ASVDD858 pKa = 3.81PTCSCVDD865 pKa = 4.36KK866 pKa = 10.74IQLGVGIGIEE876 pKa = 4.03DD877 pKa = 3.72LRR879 pKa = 11.84YY880 pKa = 10.21SADD883 pKa = 3.54GDD885 pKa = 3.85DD886 pKa = 5.36LIINFRR892 pKa = 11.84GNDD895 pKa = 3.29SDD897 pKa = 4.26SIRR900 pKa = 11.84IVGAKK905 pKa = 8.81TGASAGIEE913 pKa = 4.38SICFADD919 pKa = 4.29GTQMEE924 pKa = 4.61ISSAVTIIEE933 pKa = 4.13LTEE936 pKa = 4.13NMDD939 pKa = 3.99FQMMPVPAQTAIINGYY955 pKa = 9.08GGNDD959 pKa = 4.64FINVTGATNCIVDD972 pKa = 4.04GGSGIDD978 pKa = 3.68QIMSTAANSILYY990 pKa = 10.16GGADD994 pKa = 3.86MDD996 pKa = 4.11MLSAVNGKK1004 pKa = 8.49ATFIGGTGNDD1014 pKa = 4.07TLTGGYY1020 pKa = 10.35LGDD1023 pKa = 3.68TYY1025 pKa = 11.7VFAQGDD1031 pKa = 4.1GQDD1034 pKa = 3.98IINDD1038 pKa = 4.18NVPMTCVANFDD1049 pKa = 4.11PDD1051 pKa = 4.31TIKK1054 pKa = 10.85LGNGITKK1061 pKa = 10.35NDD1063 pKa = 2.99VAFFMEE1069 pKa = 5.16GPNLIVSYY1077 pKa = 11.26GEE1079 pKa = 3.83TDD1081 pKa = 2.99RR1082 pKa = 11.84VTVLGQANAKK1092 pKa = 9.58NAIEE1096 pKa = 4.18TVQLASGSSLSSAEE1110 pKa = 3.79INQIVADD1117 pKa = 4.46LSNYY1121 pKa = 10.36ASDD1124 pKa = 3.86HH1125 pKa = 5.98GLDD1128 pKa = 3.68FTSVEE1133 pKa = 4.14DD1134 pKa = 3.53VKK1136 pKa = 11.55NNQEE1140 pKa = 3.88LMNIVTAAWDD1150 pKa = 3.43NN1151 pKa = 3.6
GG1 pKa = 7.65EE2 pKa = 5.96DD3 pKa = 3.66GDD5 pKa = 4.42DD6 pKa = 4.12TITTTGSAYY15 pKa = 9.46QHH17 pKa = 5.19VQGGNGNDD25 pKa = 4.04TIRR28 pKa = 11.84TGSANDD34 pKa = 4.48AIIGGADD41 pKa = 4.09NDD43 pKa = 4.82TIWAGEE49 pKa = 4.09GDD51 pKa = 3.52NSIWGGTGEE60 pKa = 4.96DD61 pKa = 4.48EE62 pKa = 4.19IHH64 pKa = 6.6SGSGNDD70 pKa = 4.14TIDD73 pKa = 4.35AGDD76 pKa = 4.47DD77 pKa = 3.68NDD79 pKa = 5.1TIWAGDD85 pKa = 3.56GTNTIDD91 pKa = 3.81AGKK94 pKa = 10.76GNDD97 pKa = 4.1IIHH100 pKa = 7.0AGANADD106 pKa = 3.57TYY108 pKa = 11.02KK109 pKa = 10.58YY110 pKa = 10.95SKK112 pKa = 11.21GDD114 pKa = 3.46GHH116 pKa = 7.13DD117 pKa = 3.46VIYY120 pKa = 10.73DD121 pKa = 3.74NEE123 pKa = 4.57SGAVTADD130 pKa = 3.21HH131 pKa = 6.53TLDD134 pKa = 4.58AIRR137 pKa = 11.84FSSDD141 pKa = 2.07IAYY144 pKa = 10.51NDD146 pKa = 3.5LQFAIDD152 pKa = 4.68GNDD155 pKa = 3.6LLITFKK161 pKa = 11.0NSPEE165 pKa = 3.59DD166 pKa = 3.45SIRR169 pKa = 11.84IKK171 pKa = 10.81DD172 pKa = 3.28AATNPEE178 pKa = 4.0HH179 pKa = 6.91AVEE182 pKa = 5.47KK183 pKa = 10.6ILIEE187 pKa = 3.94NRR189 pKa = 11.84YY190 pKa = 8.97NASQVSTYY198 pKa = 9.95YY199 pKa = 10.69VEE201 pKa = 4.15NAAIRR206 pKa = 11.84HH207 pKa = 4.49TTYY210 pKa = 11.29SDD212 pKa = 3.51GDD214 pKa = 3.77DD215 pKa = 4.09TISVPVSSKK224 pKa = 8.1ITKK227 pKa = 10.13ISAGDD232 pKa = 3.66GNDD235 pKa = 3.18TVTSGDD241 pKa = 3.34SSVIIDD247 pKa = 4.1GGAGNDD253 pKa = 4.0TITTGSKK260 pKa = 10.43SDD262 pKa = 3.86VISGGTGNDD271 pKa = 3.73TINAGGGSDD280 pKa = 3.56TLTGGSGSDD289 pKa = 3.06TYY291 pKa = 11.11IFNIGDD297 pKa = 3.49GHH299 pKa = 5.95DD300 pKa = 4.03TIIEE304 pKa = 4.03ASVDD308 pKa = 3.81PTCSCVDD315 pKa = 4.07TISFGAGITSSDD327 pKa = 3.43IIYY330 pKa = 10.55KK331 pKa = 10.12VDD333 pKa = 3.53GNDD336 pKa = 4.95LIICIKK342 pKa = 10.55GHH344 pKa = 6.98DD345 pKa = 4.08GDD347 pKa = 4.67SIRR350 pKa = 11.84ITGALNGASHH360 pKa = 7.45GIEE363 pKa = 4.16TITFASGTTITLNTVISCHH382 pKa = 6.29NYY384 pKa = 8.89TDD386 pKa = 3.74ASDD389 pKa = 4.21TIVVPADD396 pKa = 3.36KK397 pKa = 10.91YY398 pKa = 10.64VVVINAGGGNDD409 pKa = 4.27TITGGDD415 pKa = 3.31TVSIINGGTGNDD427 pKa = 4.02SISTGGKK434 pKa = 10.02DD435 pKa = 3.28DD436 pKa = 3.92TLTGGEE442 pKa = 4.35GDD444 pKa = 3.72DD445 pKa = 4.49TINAGGGNDD454 pKa = 4.25TITGGTGSDD463 pKa = 3.41TLTGGSGSDD472 pKa = 3.06TYY474 pKa = 11.11IFNIGDD480 pKa = 3.49GHH482 pKa = 5.95DD483 pKa = 4.03TIIEE487 pKa = 4.03ASVDD491 pKa = 3.74PTCTCVDD498 pKa = 4.4TISFGAGITSSDD510 pKa = 3.43IIYY513 pKa = 10.55KK514 pKa = 10.12VDD516 pKa = 3.53GNDD519 pKa = 4.95LIICIKK525 pKa = 10.55GHH527 pKa = 6.98DD528 pKa = 4.08GDD530 pKa = 4.67SIRR533 pKa = 11.84ITGALNGASHH543 pKa = 7.45GIEE546 pKa = 4.16TITFASGTSITLNTVISCHH565 pKa = 6.29NYY567 pKa = 8.89TDD569 pKa = 3.74ASDD572 pKa = 4.21TIVVPADD579 pKa = 3.36KK580 pKa = 10.91YY581 pKa = 10.64VVVINAGGGNDD592 pKa = 4.27TITGGDD598 pKa = 3.31TVSIINGGTGNDD610 pKa = 4.02SISTGGKK617 pKa = 10.04DD618 pKa = 4.33DD619 pKa = 3.92ILTGGEE625 pKa = 4.02GDD627 pKa = 3.7DD628 pKa = 4.49TINAGGGNDD637 pKa = 4.13IITGGTGSDD646 pKa = 3.41TLTGGSGSDD655 pKa = 3.06TYY657 pKa = 11.11IFNIGDD663 pKa = 3.49GHH665 pKa = 5.95DD666 pKa = 4.03TIIEE670 pKa = 4.03ASVDD674 pKa = 3.81PTCSCVDD681 pKa = 4.07TISFGAGITSSDD693 pKa = 3.43IIYY696 pKa = 10.55KK697 pKa = 10.12VDD699 pKa = 3.53GNDD702 pKa = 4.95LIICIKK708 pKa = 10.55GHH710 pKa = 6.98DD711 pKa = 4.08GDD713 pKa = 4.67SIRR716 pKa = 11.84ITGALNGTSSFGVEE730 pKa = 4.15NISFTDD736 pKa = 3.87GTSIALTTVISCQNYY751 pKa = 8.51TDD753 pKa = 3.92ASDD756 pKa = 4.2TIVVPADD763 pKa = 3.34KK764 pKa = 10.77YY765 pKa = 10.03VAVINAGGGNDD776 pKa = 4.27TITGGDD782 pKa = 3.28TASIINGGAGNDD794 pKa = 4.16SISTGAKK801 pKa = 10.11DD802 pKa = 4.98DD803 pKa = 3.95ILNGGIGNDD812 pKa = 3.95TINAGGGNDD821 pKa = 4.25TITGGTGTDD830 pKa = 3.48TLIGGSGSDD839 pKa = 3.12TYY841 pKa = 11.11IFNIGDD847 pKa = 3.49GHH849 pKa = 5.95DD850 pKa = 4.03TIIEE854 pKa = 4.03ASVDD858 pKa = 3.81PTCSCVDD865 pKa = 4.36KK866 pKa = 10.74IQLGVGIGIEE876 pKa = 4.03DD877 pKa = 3.72LRR879 pKa = 11.84YY880 pKa = 10.21SADD883 pKa = 3.54GDD885 pKa = 3.85DD886 pKa = 5.36LIINFRR892 pKa = 11.84GNDD895 pKa = 3.29SDD897 pKa = 4.26SIRR900 pKa = 11.84IVGAKK905 pKa = 8.81TGASAGIEE913 pKa = 4.38SICFADD919 pKa = 4.29GTQMEE924 pKa = 4.61ISSAVTIIEE933 pKa = 4.13LTEE936 pKa = 4.13NMDD939 pKa = 3.99FQMMPVPAQTAIINGYY955 pKa = 9.08GGNDD959 pKa = 4.64FINVTGATNCIVDD972 pKa = 4.04GGSGIDD978 pKa = 3.68QIMSTAANSILYY990 pKa = 10.16GGADD994 pKa = 3.86MDD996 pKa = 4.11MLSAVNGKK1004 pKa = 8.49ATFIGGTGNDD1014 pKa = 4.07TLTGGYY1020 pKa = 10.35LGDD1023 pKa = 3.68TYY1025 pKa = 11.7VFAQGDD1031 pKa = 4.1GQDD1034 pKa = 3.98IINDD1038 pKa = 4.18NVPMTCVANFDD1049 pKa = 4.11PDD1051 pKa = 4.31TIKK1054 pKa = 10.85LGNGITKK1061 pKa = 10.35NDD1063 pKa = 2.99VAFFMEE1069 pKa = 5.16GPNLIVSYY1077 pKa = 11.26GEE1079 pKa = 3.83TDD1081 pKa = 2.99RR1082 pKa = 11.84VTVLGQANAKK1092 pKa = 9.58NAIEE1096 pKa = 4.18TVQLASGSSLSSAEE1110 pKa = 3.79INQIVADD1117 pKa = 4.46LSNYY1121 pKa = 10.36ASDD1124 pKa = 3.86HH1125 pKa = 5.98GLDD1128 pKa = 3.68FTSVEE1133 pKa = 4.14DD1134 pKa = 3.53VKK1136 pKa = 11.55NNQEE1140 pKa = 3.88LMNIVTAAWDD1150 pKa = 3.43NN1151 pKa = 3.6
Molecular weight: 116.69 kDa
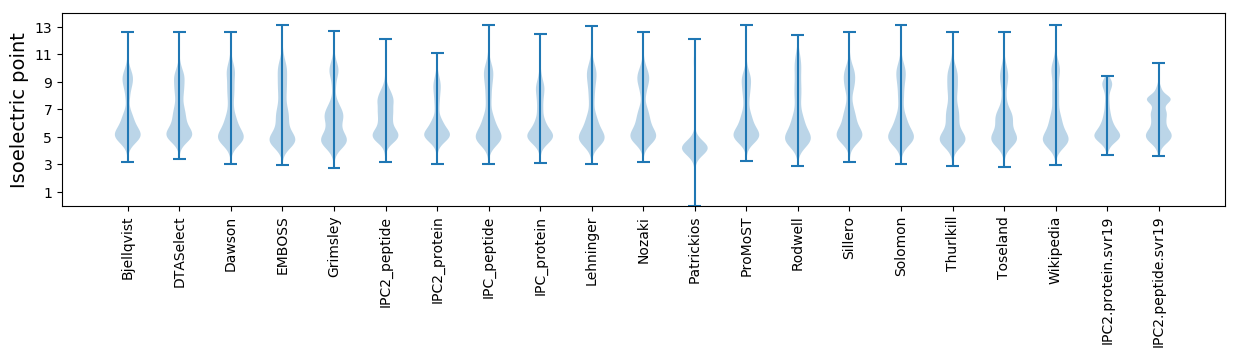
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B7XFL4|A0A1B7XFL4_9DELT tRNA modification GTPase MnmE OS=Halodesulfovibrio spirochaetisodalis OX=1560234 GN=mnmE PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.17QPSKK9 pKa = 9.44IKK11 pKa = 10.44RR12 pKa = 11.84KK13 pKa = 7.76RR14 pKa = 11.84THH16 pKa = 6.05GFRR19 pKa = 11.84ARR21 pKa = 11.84LKK23 pKa = 8.16TASGRR28 pKa = 11.84AILRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.48GRR39 pKa = 11.84AKK41 pKa = 10.69LSAA44 pKa = 3.92
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.17QPSKK9 pKa = 9.44IKK11 pKa = 10.44RR12 pKa = 11.84KK13 pKa = 7.76RR14 pKa = 11.84THH16 pKa = 6.05GFRR19 pKa = 11.84ARR21 pKa = 11.84LKK23 pKa = 8.16TASGRR28 pKa = 11.84AILRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.48GRR39 pKa = 11.84AKK41 pKa = 10.69LSAA44 pKa = 3.92
Molecular weight: 5.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
889119 |
44 |
3318 |
317.2 |
35.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.121 ± 0.048 | 1.391 ± 0.023 |
5.304 ± 0.038 | 6.752 ± 0.047 |
4.166 ± 0.034 | 7.311 ± 0.053 |
2.131 ± 0.021 | 6.282 ± 0.037 |
5.498 ± 0.046 | 9.887 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.845 ± 0.021 | 3.683 ± 0.03 |
4.194 ± 0.028 | 3.468 ± 0.03 |
4.781 ± 0.039 | 6.157 ± 0.032 |
5.775 ± 0.04 | 7.315 ± 0.04 |
1.076 ± 0.018 | 2.861 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
