
Dragonfly larvae associated circular virus-4
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 8.64
Get precalculated fractions of proteins
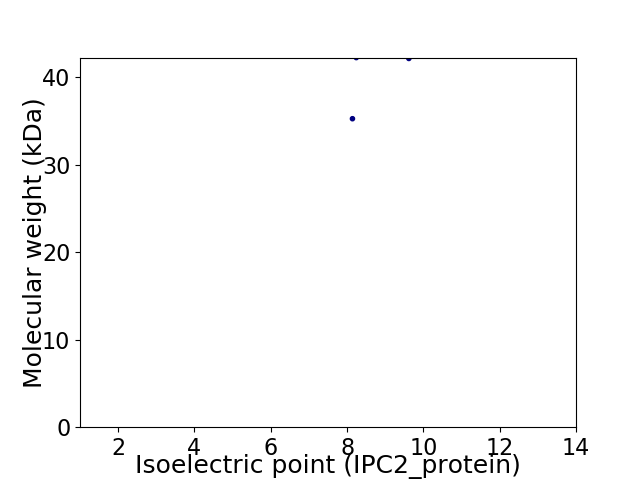
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W5U2Q0|W5U2Q0_9VIRU Putative capsid protein OS=Dragonfly larvae associated circular virus-4 OX=1454025 PE=4 SV=1
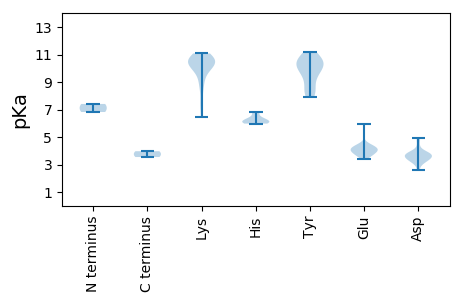
MM1 pKa = 7.41GLLRR5 pKa = 11.84VYY7 pKa = 11.04YY8 pKa = 10.14SGQQARR14 pKa = 11.84RR15 pKa = 11.84LVILAVVWPMVGTTQRR31 pKa = 11.84SRR33 pKa = 11.84GWVFTINNYY42 pKa = 9.24NEE44 pKa = 3.69WDD46 pKa = 3.76FVNISKK52 pKa = 10.88LEE54 pKa = 3.95EE55 pKa = 3.63KK56 pKa = 9.9AQYY59 pKa = 9.9YY60 pKa = 10.08IYY62 pKa = 10.5GKK64 pKa = 10.18EE65 pKa = 4.0RR66 pKa = 11.84GEE68 pKa = 4.16EE69 pKa = 4.33GTPHH73 pKa = 6.15LQGFAYY79 pKa = 9.59FKK81 pKa = 10.79QRR83 pKa = 11.84ISFNGIRR90 pKa = 11.84DD91 pKa = 3.44ILTRR95 pKa = 11.84AHH97 pKa = 6.82VEE99 pKa = 3.61IQRR102 pKa = 11.84GFNAQAIDD110 pKa = 3.65YY111 pKa = 7.88CKK113 pKa = 10.67KK114 pKa = 10.3EE115 pKa = 4.12GEE117 pKa = 4.11FTEE120 pKa = 4.03WGEE123 pKa = 4.03PSRR126 pKa = 11.84GPAGQKK132 pKa = 10.41DD133 pKa = 3.16KK134 pKa = 11.08WKK136 pKa = 10.84DD137 pKa = 3.43VLQLARR143 pKa = 11.84QGKK146 pKa = 6.44VQEE149 pKa = 3.97IEE151 pKa = 3.71EE152 pKa = 4.42RR153 pKa = 11.84YY154 pKa = 8.38PAIFLRR160 pKa = 11.84YY161 pKa = 8.2FQKK164 pKa = 10.69LCGFYY169 pKa = 10.42RR170 pKa = 11.84PEE172 pKa = 3.84HH173 pKa = 6.33SIILEE178 pKa = 4.05NFTNEE183 pKa = 3.8WWWGPTGTGKK193 pKa = 10.34FKK195 pKa = 10.94KK196 pKa = 10.75LNDD199 pKa = 4.79DD200 pKa = 3.67YY201 pKa = 11.21PDD203 pKa = 4.43PYY205 pKa = 10.81EE206 pKa = 5.29KK207 pKa = 10.81SLDD210 pKa = 3.83PWWDD214 pKa = 2.9NYY216 pKa = 8.62QRR218 pKa = 11.84EE219 pKa = 4.44EE220 pKa = 3.77IVAIEE225 pKa = 4.06EE226 pKa = 4.0FEE228 pKa = 4.24PRR230 pKa = 11.84CKK232 pKa = 10.28INSFLLKK239 pKa = 9.96RR240 pKa = 11.84WADD243 pKa = 3.87RR244 pKa = 11.84YY245 pKa = 10.21PFRR248 pKa = 11.84CEE250 pKa = 3.5VKK252 pKa = 10.51GAFLSKK258 pKa = 10.34LRR260 pKa = 11.84PLKK263 pKa = 10.55IIVISNYY270 pKa = 9.75QLDD273 pKa = 3.75EE274 pKa = 4.42CFPNSKK280 pKa = 10.59DD281 pKa = 3.48LDD283 pKa = 3.79PLKK286 pKa = 10.81RR287 pKa = 11.84RR288 pKa = 11.84FKK290 pKa = 10.35EE291 pKa = 3.4IHH293 pKa = 6.05FPP295 pKa = 3.97
MM1 pKa = 7.41GLLRR5 pKa = 11.84VYY7 pKa = 11.04YY8 pKa = 10.14SGQQARR14 pKa = 11.84RR15 pKa = 11.84LVILAVVWPMVGTTQRR31 pKa = 11.84SRR33 pKa = 11.84GWVFTINNYY42 pKa = 9.24NEE44 pKa = 3.69WDD46 pKa = 3.76FVNISKK52 pKa = 10.88LEE54 pKa = 3.95EE55 pKa = 3.63KK56 pKa = 9.9AQYY59 pKa = 9.9YY60 pKa = 10.08IYY62 pKa = 10.5GKK64 pKa = 10.18EE65 pKa = 4.0RR66 pKa = 11.84GEE68 pKa = 4.16EE69 pKa = 4.33GTPHH73 pKa = 6.15LQGFAYY79 pKa = 9.59FKK81 pKa = 10.79QRR83 pKa = 11.84ISFNGIRR90 pKa = 11.84DD91 pKa = 3.44ILTRR95 pKa = 11.84AHH97 pKa = 6.82VEE99 pKa = 3.61IQRR102 pKa = 11.84GFNAQAIDD110 pKa = 3.65YY111 pKa = 7.88CKK113 pKa = 10.67KK114 pKa = 10.3EE115 pKa = 4.12GEE117 pKa = 4.11FTEE120 pKa = 4.03WGEE123 pKa = 4.03PSRR126 pKa = 11.84GPAGQKK132 pKa = 10.41DD133 pKa = 3.16KK134 pKa = 11.08WKK136 pKa = 10.84DD137 pKa = 3.43VLQLARR143 pKa = 11.84QGKK146 pKa = 6.44VQEE149 pKa = 3.97IEE151 pKa = 3.71EE152 pKa = 4.42RR153 pKa = 11.84YY154 pKa = 8.38PAIFLRR160 pKa = 11.84YY161 pKa = 8.2FQKK164 pKa = 10.69LCGFYY169 pKa = 10.42RR170 pKa = 11.84PEE172 pKa = 3.84HH173 pKa = 6.33SIILEE178 pKa = 4.05NFTNEE183 pKa = 3.8WWWGPTGTGKK193 pKa = 10.34FKK195 pKa = 10.94KK196 pKa = 10.75LNDD199 pKa = 4.79DD200 pKa = 3.67YY201 pKa = 11.21PDD203 pKa = 4.43PYY205 pKa = 10.81EE206 pKa = 5.29KK207 pKa = 10.81SLDD210 pKa = 3.83PWWDD214 pKa = 2.9NYY216 pKa = 8.62QRR218 pKa = 11.84EE219 pKa = 4.44EE220 pKa = 3.77IVAIEE225 pKa = 4.06EE226 pKa = 4.0FEE228 pKa = 4.24PRR230 pKa = 11.84CKK232 pKa = 10.28INSFLLKK239 pKa = 9.96RR240 pKa = 11.84WADD243 pKa = 3.87RR244 pKa = 11.84YY245 pKa = 10.21PFRR248 pKa = 11.84CEE250 pKa = 3.5VKK252 pKa = 10.51GAFLSKK258 pKa = 10.34LRR260 pKa = 11.84PLKK263 pKa = 10.55IIVISNYY270 pKa = 9.75QLDD273 pKa = 3.75EE274 pKa = 4.42CFPNSKK280 pKa = 10.59DD281 pKa = 3.48LDD283 pKa = 3.79PLKK286 pKa = 10.81RR287 pKa = 11.84RR288 pKa = 11.84FKK290 pKa = 10.35EE291 pKa = 3.4IHH293 pKa = 6.05FPP295 pKa = 3.97
Molecular weight: 35.24 kDa
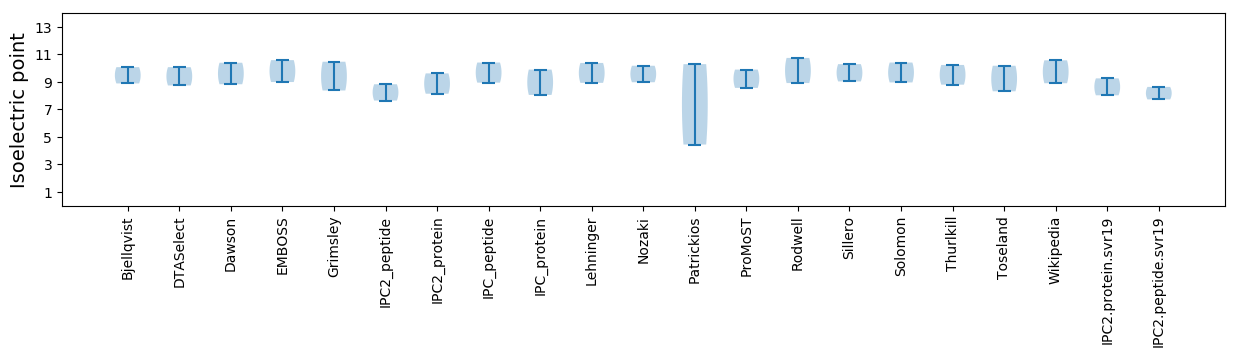
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W5U2Q0|W5U2Q0_9VIRU Putative capsid protein OS=Dragonfly larvae associated circular virus-4 OX=1454025 PE=4 SV=1
MM1 pKa = 6.82YY2 pKa = 10.24GKK4 pKa = 10.17RR5 pKa = 11.84KK6 pKa = 7.12YY7 pKa = 10.78AKK9 pKa = 9.09YY10 pKa = 10.37ASAFAFRR17 pKa = 11.84RR18 pKa = 11.84NMRR21 pKa = 11.84TRR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84YY26 pKa = 8.61GAPGDD31 pKa = 3.8PSMTSIGNISPTGRR45 pKa = 11.84SIARR49 pKa = 11.84ANRR52 pKa = 11.84SNIGRR57 pKa = 11.84TLTGTTRR64 pKa = 11.84SRR66 pKa = 11.84YY67 pKa = 9.71NSGQGVTQQHH77 pKa = 6.14DD78 pKa = 3.32VRR80 pKa = 11.84LIYY83 pKa = 10.3AKK85 pKa = 10.52KK86 pKa = 10.43RR87 pKa = 11.84MPNYY91 pKa = 9.66KK92 pKa = 9.7KK93 pKa = 10.5KK94 pKa = 8.51PWRR97 pKa = 11.84RR98 pKa = 11.84FVKK101 pKa = 10.14KK102 pKa = 8.76VHH104 pKa = 5.98AVSEE108 pKa = 4.21KK109 pKa = 10.99DD110 pKa = 3.22LGSRR114 pKa = 11.84TALFSNTITYY124 pKa = 10.77NNTVDD129 pKa = 4.94GADD132 pKa = 3.28EE133 pKa = 4.42CLTIGLYY140 pKa = 9.81PLQSTDD146 pKa = 2.59PWLNDD151 pKa = 3.15LNTITSLEE159 pKa = 4.15NQGNPTAAAGGTVDD173 pKa = 4.08PTTKK177 pKa = 10.46FIFQSGVLDD186 pKa = 3.41LTIRR190 pKa = 11.84NATTQTGQLNPDD202 pKa = 3.65QFNAAATQEE211 pKa = 3.7IDD213 pKa = 3.49IYY215 pKa = 11.03EE216 pKa = 4.19IMCRR220 pKa = 11.84ADD222 pKa = 3.84FMQVSGGNKK231 pKa = 9.26INLSQCFEE239 pKa = 4.37EE240 pKa = 5.91GSVITRR246 pKa = 11.84NIANAPGDD254 pKa = 3.52IKK256 pKa = 10.61IANRR260 pKa = 11.84GATPFDD266 pKa = 3.83LPAALSRR273 pKa = 11.84YY274 pKa = 8.13GMKK277 pKa = 9.94IIKK280 pKa = 7.49KK281 pKa = 7.63TKK283 pKa = 8.73YY284 pKa = 9.06FVPGGGTITYY294 pKa = 9.12QIRR297 pKa = 11.84DD298 pKa = 3.64PKK300 pKa = 10.69RR301 pKa = 11.84RR302 pKa = 11.84VSSFRR307 pKa = 11.84DD308 pKa = 3.38LNVEE312 pKa = 3.83GGVNKK317 pKa = 10.2VGWTKK322 pKa = 11.01YY323 pKa = 10.64LYY325 pKa = 10.33INAKK329 pKa = 9.41VIPGFLLGTGPASFVQRR346 pKa = 11.84MQVGVTRR353 pKa = 11.84KK354 pKa = 9.96YY355 pKa = 8.0MYY357 pKa = 10.27KK358 pKa = 9.92VEE360 pKa = 4.35GFNEE364 pKa = 4.13SRR366 pKa = 11.84DD367 pKa = 3.74SYY369 pKa = 10.82STLNSTISAPAA380 pKa = 3.55
MM1 pKa = 6.82YY2 pKa = 10.24GKK4 pKa = 10.17RR5 pKa = 11.84KK6 pKa = 7.12YY7 pKa = 10.78AKK9 pKa = 9.09YY10 pKa = 10.37ASAFAFRR17 pKa = 11.84RR18 pKa = 11.84NMRR21 pKa = 11.84TRR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84YY26 pKa = 8.61GAPGDD31 pKa = 3.8PSMTSIGNISPTGRR45 pKa = 11.84SIARR49 pKa = 11.84ANRR52 pKa = 11.84SNIGRR57 pKa = 11.84TLTGTTRR64 pKa = 11.84SRR66 pKa = 11.84YY67 pKa = 9.71NSGQGVTQQHH77 pKa = 6.14DD78 pKa = 3.32VRR80 pKa = 11.84LIYY83 pKa = 10.3AKK85 pKa = 10.52KK86 pKa = 10.43RR87 pKa = 11.84MPNYY91 pKa = 9.66KK92 pKa = 9.7KK93 pKa = 10.5KK94 pKa = 8.51PWRR97 pKa = 11.84RR98 pKa = 11.84FVKK101 pKa = 10.14KK102 pKa = 8.76VHH104 pKa = 5.98AVSEE108 pKa = 4.21KK109 pKa = 10.99DD110 pKa = 3.22LGSRR114 pKa = 11.84TALFSNTITYY124 pKa = 10.77NNTVDD129 pKa = 4.94GADD132 pKa = 3.28EE133 pKa = 4.42CLTIGLYY140 pKa = 9.81PLQSTDD146 pKa = 2.59PWLNDD151 pKa = 3.15LNTITSLEE159 pKa = 4.15NQGNPTAAAGGTVDD173 pKa = 4.08PTTKK177 pKa = 10.46FIFQSGVLDD186 pKa = 3.41LTIRR190 pKa = 11.84NATTQTGQLNPDD202 pKa = 3.65QFNAAATQEE211 pKa = 3.7IDD213 pKa = 3.49IYY215 pKa = 11.03EE216 pKa = 4.19IMCRR220 pKa = 11.84ADD222 pKa = 3.84FMQVSGGNKK231 pKa = 9.26INLSQCFEE239 pKa = 4.37EE240 pKa = 5.91GSVITRR246 pKa = 11.84NIANAPGDD254 pKa = 3.52IKK256 pKa = 10.61IANRR260 pKa = 11.84GATPFDD266 pKa = 3.83LPAALSRR273 pKa = 11.84YY274 pKa = 8.13GMKK277 pKa = 9.94IIKK280 pKa = 7.49KK281 pKa = 7.63TKK283 pKa = 8.73YY284 pKa = 9.06FVPGGGTITYY294 pKa = 9.12QIRR297 pKa = 11.84DD298 pKa = 3.64PKK300 pKa = 10.69RR301 pKa = 11.84RR302 pKa = 11.84VSSFRR307 pKa = 11.84DD308 pKa = 3.38LNVEE312 pKa = 3.83GGVNKK317 pKa = 10.2VGWTKK322 pKa = 11.01YY323 pKa = 10.64LYY325 pKa = 10.33INAKK329 pKa = 9.41VIPGFLLGTGPASFVQRR346 pKa = 11.84MQVGVTRR353 pKa = 11.84KK354 pKa = 9.96YY355 pKa = 8.0MYY357 pKa = 10.27KK358 pKa = 9.92VEE360 pKa = 4.35GFNEE364 pKa = 4.13SRR366 pKa = 11.84DD367 pKa = 3.74SYY369 pKa = 10.82STLNSTISAPAA380 pKa = 3.55
Molecular weight: 42.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
675 |
295 |
380 |
337.5 |
38.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.222 ± 1.137 | 1.185 ± 0.319 |
4.593 ± 0.096 | 5.333 ± 2.18 |
5.037 ± 0.879 | 8.0 ± 0.764 |
0.889 ± 0.293 | 6.667 ± 0.071 |
6.963 ± 0.522 | 6.37 ± 0.681 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.63 ± 0.596 | 5.778 ± 0.859 |
5.037 ± 0.242 | 4.444 ± 0.401 |
7.852 ± 0.035 | 5.481 ± 1.098 |
6.519 ± 2.172 | 4.889 ± 0.302 |
2.074 ± 1.036 | 5.037 ± 0.242 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
