
Clostridium bartlettii CAG:1329
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Clostridiaceae; Clostridium; environmental samples
Average proteome isoelectric point is 6.27
Get precalculated fractions of proteins
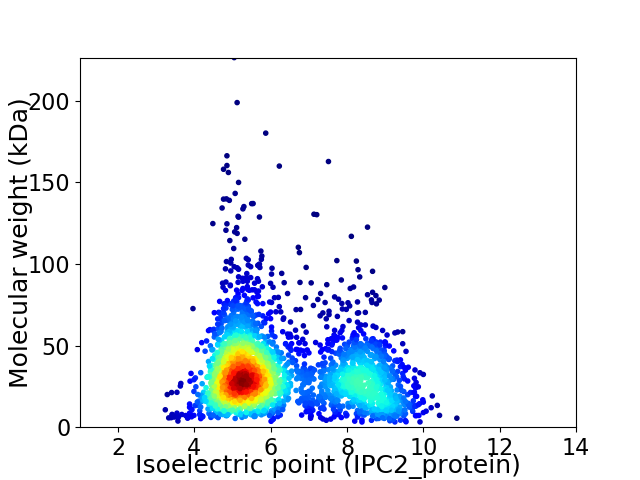
Virtual 2D-PAGE plot for 2686 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R5XNK5|R5XNK5_9CLOT Lar_N domain-containing protein OS=Clostridium bartlettii CAG:1329 OX=1263063 GN=BN488_01405 PE=4 SV=1
MM1 pKa = 7.96RR2 pKa = 11.84EE3 pKa = 3.72ATLPFTVTKK12 pKa = 10.47SVNPTSAGLNDD23 pKa = 3.65TVTYY27 pKa = 9.45TVVLNNTSANTLSNATFVDD46 pKa = 3.72QLPAGLEE53 pKa = 4.12YY54 pKa = 11.01VPNTLTVNGQAKK66 pKa = 10.46AGVDD70 pKa = 3.57PNTQFTIDD78 pKa = 5.02DD79 pKa = 3.59IPGNGNATVTFDD91 pKa = 3.72AKK93 pKa = 10.77VVSLPATPKK102 pKa = 10.1TEE104 pKa = 3.75NTAQFGFSYY113 pKa = 10.45TPITGSPAQTLNLSSTEE130 pKa = 3.94ANPLYY135 pKa = 10.52IGDD138 pKa = 4.14AVIGSTNFTKK148 pKa = 10.39TAGATSAKK156 pKa = 10.27VGDD159 pKa = 4.53EE160 pKa = 3.87IPYY163 pKa = 9.58TIAISNGGNVNATNVTLTDD182 pKa = 3.99ALPPGTTLVADD193 pKa = 4.06SLTVTNKK200 pKa = 7.36QTGANVDD207 pKa = 3.63YY208 pKa = 10.77TGDD211 pKa = 3.69LASGLNLTNALQPGTANEE229 pKa = 3.87VDD231 pKa = 3.35VSYY234 pKa = 10.98RR235 pKa = 11.84ALVNTAPVGNTLTNTANLSYY255 pKa = 10.48QYY257 pKa = 10.76QRR259 pKa = 11.84NPNDD263 pKa = 3.57PTTVTQTATSSANVNIEE280 pKa = 3.8NASDD284 pKa = 3.73YY285 pKa = 10.75PITITKK291 pKa = 7.62TANPANNLVVGDD303 pKa = 3.91VVTYY307 pKa = 9.08TVTVEE312 pKa = 3.92NTSTNNATYY321 pKa = 10.3TNTTITDD328 pKa = 3.91TLPDD332 pKa = 3.45NLTYY336 pKa = 10.73EE337 pKa = 4.44RR338 pKa = 11.84GSVTINGQTSTQEE351 pKa = 3.83LTNGITLEE359 pKa = 4.18NDD361 pKa = 3.74LAPNVPTTVTFRR373 pKa = 11.84AVVNSIPQNKK383 pKa = 8.47TISNTASINYY393 pKa = 7.8DD394 pKa = 3.06ATVGGEE400 pKa = 3.92IKK402 pKa = 10.19QQNIDD407 pKa = 3.57ATSEE411 pKa = 4.44AITSNVVNASLNVTKK426 pKa = 10.24TDD428 pKa = 3.33NAAFTLAIGDD438 pKa = 3.96TFNYY442 pKa = 8.31TISVSNTGEE451 pKa = 3.98VPLNNVTVTDD461 pKa = 4.01NLPSQLEE468 pKa = 4.25VTQISVDD475 pKa = 3.55GAPIDD480 pKa = 3.73TLLTNNIPLGDD491 pKa = 3.74MNANEE496 pKa = 4.44TKK498 pKa = 10.24TVVISVKK505 pKa = 10.3AASGGLNNFQNTAIASSQVNIEE527 pKa = 3.97GRR529 pKa = 11.84TEE531 pKa = 3.58PAEE534 pKa = 3.97ISISGSDD541 pKa = 3.46NNAITIPTNPTPIIPTPTPNLLNLASINSIASKK574 pKa = 10.18NISYY578 pKa = 10.79VGDD581 pKa = 4.55AISYY585 pKa = 7.71ATIVSNVGNVPLGTYY600 pKa = 9.36DD601 pKa = 3.39VPVTVFTEE609 pKa = 4.25LSPSVSFIPGSLSINNVPVNCVNPSQINLGILQPGDD645 pKa = 3.41VRR647 pKa = 11.84IITYY651 pKa = 7.75SVRR654 pKa = 11.84VNNACPNIVYY664 pKa = 10.38NIQKK668 pKa = 10.17LLYY671 pKa = 9.75GYY673 pKa = 10.29DD674 pKa = 3.48PLYY677 pKa = 10.59IGTSTFSVNGNIVMTYY693 pKa = 10.38INN695 pKa = 3.81
MM1 pKa = 7.96RR2 pKa = 11.84EE3 pKa = 3.72ATLPFTVTKK12 pKa = 10.47SVNPTSAGLNDD23 pKa = 3.65TVTYY27 pKa = 9.45TVVLNNTSANTLSNATFVDD46 pKa = 3.72QLPAGLEE53 pKa = 4.12YY54 pKa = 11.01VPNTLTVNGQAKK66 pKa = 10.46AGVDD70 pKa = 3.57PNTQFTIDD78 pKa = 5.02DD79 pKa = 3.59IPGNGNATVTFDD91 pKa = 3.72AKK93 pKa = 10.77VVSLPATPKK102 pKa = 10.1TEE104 pKa = 3.75NTAQFGFSYY113 pKa = 10.45TPITGSPAQTLNLSSTEE130 pKa = 3.94ANPLYY135 pKa = 10.52IGDD138 pKa = 4.14AVIGSTNFTKK148 pKa = 10.39TAGATSAKK156 pKa = 10.27VGDD159 pKa = 4.53EE160 pKa = 3.87IPYY163 pKa = 9.58TIAISNGGNVNATNVTLTDD182 pKa = 3.99ALPPGTTLVADD193 pKa = 4.06SLTVTNKK200 pKa = 7.36QTGANVDD207 pKa = 3.63YY208 pKa = 10.77TGDD211 pKa = 3.69LASGLNLTNALQPGTANEE229 pKa = 3.87VDD231 pKa = 3.35VSYY234 pKa = 10.98RR235 pKa = 11.84ALVNTAPVGNTLTNTANLSYY255 pKa = 10.48QYY257 pKa = 10.76QRR259 pKa = 11.84NPNDD263 pKa = 3.57PTTVTQTATSSANVNIEE280 pKa = 3.8NASDD284 pKa = 3.73YY285 pKa = 10.75PITITKK291 pKa = 7.62TANPANNLVVGDD303 pKa = 3.91VVTYY307 pKa = 9.08TVTVEE312 pKa = 3.92NTSTNNATYY321 pKa = 10.3TNTTITDD328 pKa = 3.91TLPDD332 pKa = 3.45NLTYY336 pKa = 10.73EE337 pKa = 4.44RR338 pKa = 11.84GSVTINGQTSTQEE351 pKa = 3.83LTNGITLEE359 pKa = 4.18NDD361 pKa = 3.74LAPNVPTTVTFRR373 pKa = 11.84AVVNSIPQNKK383 pKa = 8.47TISNTASINYY393 pKa = 7.8DD394 pKa = 3.06ATVGGEE400 pKa = 3.92IKK402 pKa = 10.19QQNIDD407 pKa = 3.57ATSEE411 pKa = 4.44AITSNVVNASLNVTKK426 pKa = 10.24TDD428 pKa = 3.33NAAFTLAIGDD438 pKa = 3.96TFNYY442 pKa = 8.31TISVSNTGEE451 pKa = 3.98VPLNNVTVTDD461 pKa = 4.01NLPSQLEE468 pKa = 4.25VTQISVDD475 pKa = 3.55GAPIDD480 pKa = 3.73TLLTNNIPLGDD491 pKa = 3.74MNANEE496 pKa = 4.44TKK498 pKa = 10.24TVVISVKK505 pKa = 10.3AASGGLNNFQNTAIASSQVNIEE527 pKa = 3.97GRR529 pKa = 11.84TEE531 pKa = 3.58PAEE534 pKa = 3.97ISISGSDD541 pKa = 3.46NNAITIPTNPTPIIPTPTPNLLNLASINSIASKK574 pKa = 10.18NISYY578 pKa = 10.79VGDD581 pKa = 4.55AISYY585 pKa = 7.71ATIVSNVGNVPLGTYY600 pKa = 9.36DD601 pKa = 3.39VPVTVFTEE609 pKa = 4.25LSPSVSFIPGSLSINNVPVNCVNPSQINLGILQPGDD645 pKa = 3.41VRR647 pKa = 11.84IITYY651 pKa = 7.75SVRR654 pKa = 11.84VNNACPNIVYY664 pKa = 10.38NIQKK668 pKa = 10.17LLYY671 pKa = 9.75GYY673 pKa = 10.29DD674 pKa = 3.48PLYY677 pKa = 10.59IGTSTFSVNGNIVMTYY693 pKa = 10.38INN695 pKa = 3.81
Molecular weight: 72.62 kDa
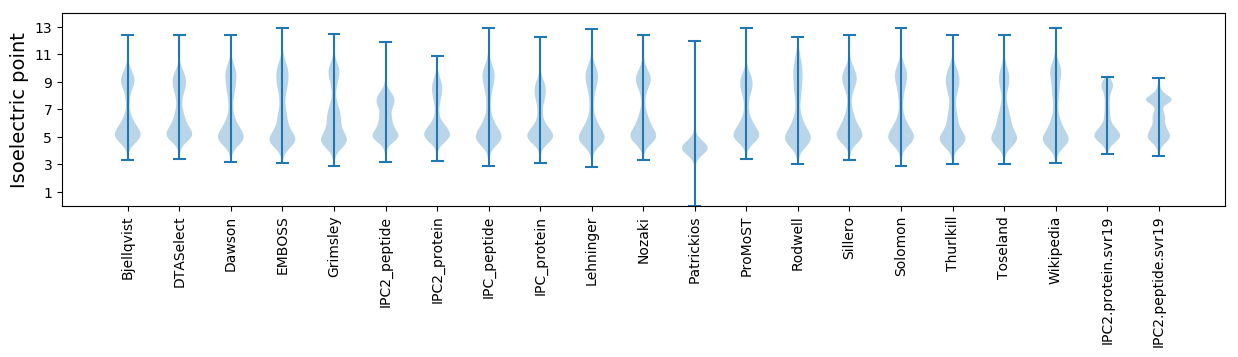
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R5XGS9|R5XGS9_9CLOT Ion-translocating oxidoreductase complex subunit D OS=Clostridium bartlettii CAG:1329 OX=1263063 GN=rnfD PE=3 SV=1
MM1 pKa = 7.36KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.67QPKK8 pKa = 9.0KK9 pKa = 7.87RR10 pKa = 11.84QRR12 pKa = 11.84KK13 pKa = 8.39KK14 pKa = 8.49EE15 pKa = 3.5HH16 pKa = 6.11GFRR19 pKa = 11.84KK20 pKa = 9.77RR21 pKa = 11.84MKK23 pKa = 9.11TSNGRR28 pKa = 11.84NVLKK32 pKa = 10.51RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.9GRR39 pKa = 11.84NRR41 pKa = 11.84LTHH44 pKa = 6.18
MM1 pKa = 7.36KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.67QPKK8 pKa = 9.0KK9 pKa = 7.87RR10 pKa = 11.84QRR12 pKa = 11.84KK13 pKa = 8.39KK14 pKa = 8.49EE15 pKa = 3.5HH16 pKa = 6.11GFRR19 pKa = 11.84KK20 pKa = 9.77RR21 pKa = 11.84MKK23 pKa = 9.11TSNGRR28 pKa = 11.84NVLKK32 pKa = 10.51RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.9GRR39 pKa = 11.84NRR41 pKa = 11.84LTHH44 pKa = 6.18
Molecular weight: 5.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
839455 |
29 |
2011 |
312.5 |
35.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.61 ± 0.049 | 1.361 ± 0.022 |
6.12 ± 0.046 | 7.36 ± 0.058 |
4.237 ± 0.034 | 6.313 ± 0.051 |
1.34 ± 0.018 | 9.621 ± 0.058 |
9.131 ± 0.057 | 8.85 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.682 ± 0.023 | 6.319 ± 0.048 |
2.684 ± 0.027 | 2.567 ± 0.023 |
3.177 ± 0.03 | 6.112 ± 0.042 |
5.023 ± 0.04 | 6.627 ± 0.043 |
0.581 ± 0.012 | 4.285 ± 0.037 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
