
Xinzhou dimarhabdovirus virus 1
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.27
Get precalculated fractions of proteins
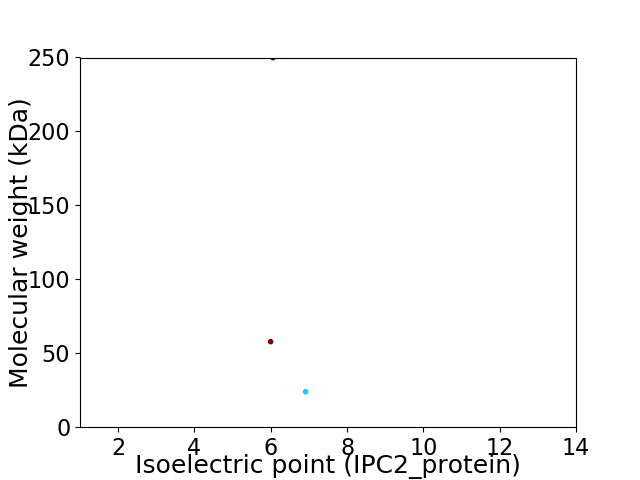
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KND6|A0A1L3KND6_9VIRU Putative glycoprotein OS=Xinzhou dimarhabdovirus virus 1 OX=1923768 PE=4 SV=1
MM1 pKa = 7.29LTLISTVLFLLSSKK15 pKa = 10.26FVLGGLIIPLCRR27 pKa = 11.84SGPLWHH33 pKa = 7.31KK34 pKa = 10.72INPTDD39 pKa = 3.96MICPRR44 pKa = 11.84YY45 pKa = 9.74EE46 pKa = 3.97SGPPEE51 pKa = 4.71GEE53 pKa = 3.9MNMTDD58 pKa = 2.98IQTFDD63 pKa = 3.18IPAISGKK70 pKa = 8.38QLGYY74 pKa = 9.88LCHH77 pKa = 5.88GVIYY81 pKa = 10.14RR82 pKa = 11.84VICIKK87 pKa = 10.78GVFGGEE93 pKa = 4.33TINKK97 pKa = 9.2VILPTKK103 pKa = 9.21ITPNEE108 pKa = 3.92CEE110 pKa = 4.3VAVKK114 pKa = 10.26EE115 pKa = 4.13YY116 pKa = 9.89LTTAEE121 pKa = 4.53DD122 pKa = 3.43HH123 pKa = 6.71RR124 pKa = 11.84SGYY127 pKa = 10.26FPGKK131 pKa = 8.0YY132 pKa = 8.9CVYY135 pKa = 9.97EE136 pKa = 3.82ILSTHH141 pKa = 6.0TEE143 pKa = 3.81FSDD146 pKa = 3.33KK147 pKa = 11.01KK148 pKa = 10.11FILVSDD154 pKa = 3.73HH155 pKa = 6.05TVLYY159 pKa = 10.53DD160 pKa = 3.79PYY162 pKa = 10.59ADD164 pKa = 3.39QWIDD168 pKa = 3.34TLFLGGRR175 pKa = 11.84CDD177 pKa = 3.5TRR179 pKa = 11.84SCRR182 pKa = 11.84TIKK185 pKa = 10.86DD186 pKa = 3.55SVVWISNVVKK196 pKa = 9.29PACPKK201 pKa = 9.99AVSLLVHH208 pKa = 6.02VAYY211 pKa = 10.09EE212 pKa = 4.36GKK214 pKa = 10.35KK215 pKa = 9.48PLTIHH220 pKa = 6.8GPTIPTSKK228 pKa = 11.05LEE230 pKa = 4.24GACVTKK236 pKa = 10.65FCGTKK241 pKa = 10.29GLRR244 pKa = 11.84LSSGFFIIPPASWIKK259 pKa = 10.42VFKK262 pKa = 9.99TDD264 pKa = 3.34CTDD267 pKa = 3.06GVSIRR272 pKa = 11.84GITWEE277 pKa = 4.44SVVQDD282 pKa = 3.03TVMYY286 pKa = 10.84NEE288 pKa = 4.14ISVIRR293 pKa = 11.84LHH295 pKa = 6.58CLNTIATMSALGRR308 pKa = 11.84ASSVQLGIFQPWTPGIHH325 pKa = 5.33PVYY328 pKa = 10.28RR329 pKa = 11.84VGVDD333 pKa = 3.04NHH335 pKa = 7.18LEE337 pKa = 4.2TKK339 pKa = 8.31MCGYY343 pKa = 10.24ILGEE347 pKa = 4.43GIDD350 pKa = 3.9MNEE353 pKa = 4.54DD354 pKa = 3.02GAIGKK359 pKa = 8.75DD360 pKa = 3.49HH361 pKa = 7.27LGHH364 pKa = 6.94LVFWQDD370 pKa = 2.46WVSPRR375 pKa = 11.84KK376 pKa = 9.72LSCEE380 pKa = 3.71AYY382 pKa = 9.8GPNGIYY388 pKa = 9.78RR389 pKa = 11.84DD390 pKa = 4.06EE391 pKa = 4.47KK392 pKa = 10.89CQLHH396 pKa = 5.66YY397 pKa = 10.4PWIEE401 pKa = 3.62LRR403 pKa = 11.84QDD405 pKa = 2.61IMHH408 pKa = 7.02RR409 pKa = 11.84SILVQQTLQEE419 pKa = 4.4APHH422 pKa = 5.71VTEE425 pKa = 3.51QHH427 pKa = 5.51YY428 pKa = 10.81SQTEE432 pKa = 4.21LEE434 pKa = 4.41SDD436 pKa = 3.61FQLFDD441 pKa = 4.81KK442 pKa = 11.05DD443 pKa = 3.69FTDD446 pKa = 3.58TKK448 pKa = 10.08DD449 pKa = 2.93IEE451 pKa = 4.22MAIFSWTSSVRR462 pKa = 11.84LGLAIGGVVLVIIVGLVLLVKK483 pKa = 10.51LGLIAYY489 pKa = 8.59IWDD492 pKa = 3.38KK493 pKa = 10.56CCRR496 pKa = 11.84RR497 pKa = 11.84KK498 pKa = 8.0QHH500 pKa = 5.54RR501 pKa = 11.84QEE503 pKa = 4.71SIHH506 pKa = 6.88LEE508 pKa = 4.06PLNPIYY514 pKa = 10.88SS515 pKa = 3.6
MM1 pKa = 7.29LTLISTVLFLLSSKK15 pKa = 10.26FVLGGLIIPLCRR27 pKa = 11.84SGPLWHH33 pKa = 7.31KK34 pKa = 10.72INPTDD39 pKa = 3.96MICPRR44 pKa = 11.84YY45 pKa = 9.74EE46 pKa = 3.97SGPPEE51 pKa = 4.71GEE53 pKa = 3.9MNMTDD58 pKa = 2.98IQTFDD63 pKa = 3.18IPAISGKK70 pKa = 8.38QLGYY74 pKa = 9.88LCHH77 pKa = 5.88GVIYY81 pKa = 10.14RR82 pKa = 11.84VICIKK87 pKa = 10.78GVFGGEE93 pKa = 4.33TINKK97 pKa = 9.2VILPTKK103 pKa = 9.21ITPNEE108 pKa = 3.92CEE110 pKa = 4.3VAVKK114 pKa = 10.26EE115 pKa = 4.13YY116 pKa = 9.89LTTAEE121 pKa = 4.53DD122 pKa = 3.43HH123 pKa = 6.71RR124 pKa = 11.84SGYY127 pKa = 10.26FPGKK131 pKa = 8.0YY132 pKa = 8.9CVYY135 pKa = 9.97EE136 pKa = 3.82ILSTHH141 pKa = 6.0TEE143 pKa = 3.81FSDD146 pKa = 3.33KK147 pKa = 11.01KK148 pKa = 10.11FILVSDD154 pKa = 3.73HH155 pKa = 6.05TVLYY159 pKa = 10.53DD160 pKa = 3.79PYY162 pKa = 10.59ADD164 pKa = 3.39QWIDD168 pKa = 3.34TLFLGGRR175 pKa = 11.84CDD177 pKa = 3.5TRR179 pKa = 11.84SCRR182 pKa = 11.84TIKK185 pKa = 10.86DD186 pKa = 3.55SVVWISNVVKK196 pKa = 9.29PACPKK201 pKa = 9.99AVSLLVHH208 pKa = 6.02VAYY211 pKa = 10.09EE212 pKa = 4.36GKK214 pKa = 10.35KK215 pKa = 9.48PLTIHH220 pKa = 6.8GPTIPTSKK228 pKa = 11.05LEE230 pKa = 4.24GACVTKK236 pKa = 10.65FCGTKK241 pKa = 10.29GLRR244 pKa = 11.84LSSGFFIIPPASWIKK259 pKa = 10.42VFKK262 pKa = 9.99TDD264 pKa = 3.34CTDD267 pKa = 3.06GVSIRR272 pKa = 11.84GITWEE277 pKa = 4.44SVVQDD282 pKa = 3.03TVMYY286 pKa = 10.84NEE288 pKa = 4.14ISVIRR293 pKa = 11.84LHH295 pKa = 6.58CLNTIATMSALGRR308 pKa = 11.84ASSVQLGIFQPWTPGIHH325 pKa = 5.33PVYY328 pKa = 10.28RR329 pKa = 11.84VGVDD333 pKa = 3.04NHH335 pKa = 7.18LEE337 pKa = 4.2TKK339 pKa = 8.31MCGYY343 pKa = 10.24ILGEE347 pKa = 4.43GIDD350 pKa = 3.9MNEE353 pKa = 4.54DD354 pKa = 3.02GAIGKK359 pKa = 8.75DD360 pKa = 3.49HH361 pKa = 7.27LGHH364 pKa = 6.94LVFWQDD370 pKa = 2.46WVSPRR375 pKa = 11.84KK376 pKa = 9.72LSCEE380 pKa = 3.71AYY382 pKa = 9.8GPNGIYY388 pKa = 9.78RR389 pKa = 11.84DD390 pKa = 4.06EE391 pKa = 4.47KK392 pKa = 10.89CQLHH396 pKa = 5.66YY397 pKa = 10.4PWIEE401 pKa = 3.62LRR403 pKa = 11.84QDD405 pKa = 2.61IMHH408 pKa = 7.02RR409 pKa = 11.84SILVQQTLQEE419 pKa = 4.4APHH422 pKa = 5.71VTEE425 pKa = 3.51QHH427 pKa = 5.51YY428 pKa = 10.81SQTEE432 pKa = 4.21LEE434 pKa = 4.41SDD436 pKa = 3.61FQLFDD441 pKa = 4.81KK442 pKa = 11.05DD443 pKa = 3.69FTDD446 pKa = 3.58TKK448 pKa = 10.08DD449 pKa = 2.93IEE451 pKa = 4.22MAIFSWTSSVRR462 pKa = 11.84LGLAIGGVVLVIIVGLVLLVKK483 pKa = 10.51LGLIAYY489 pKa = 8.59IWDD492 pKa = 3.38KK493 pKa = 10.56CCRR496 pKa = 11.84RR497 pKa = 11.84KK498 pKa = 8.0QHH500 pKa = 5.54RR501 pKa = 11.84QEE503 pKa = 4.71SIHH506 pKa = 6.88LEE508 pKa = 4.06PLNPIYY514 pKa = 10.88SS515 pKa = 3.6
Molecular weight: 57.83 kDa
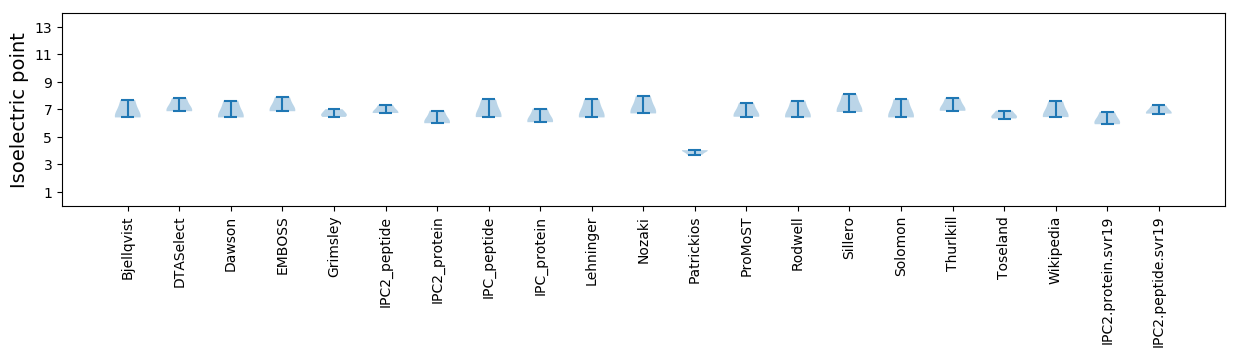
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KND6|A0A1L3KND6_9VIRU Putative glycoprotein OS=Xinzhou dimarhabdovirus virus 1 OX=1923768 PE=4 SV=1
MM1 pKa = 7.34KK2 pKa = 10.42RR3 pKa = 11.84ILSKK7 pKa = 10.64SWWTSKK13 pKa = 9.9SHH15 pKa = 4.26SHH17 pKa = 6.07EE18 pKa = 4.19MGEE21 pKa = 4.5SSGSHH26 pKa = 6.97PEE28 pKa = 4.12SQSVPVSWDD37 pKa = 2.84QGVALTEE44 pKa = 4.35PPSAPLSSSGPFCLDD59 pKa = 3.58LEE61 pKa = 4.71VEE63 pKa = 4.72AYY65 pKa = 10.82LHH67 pKa = 5.23VQCNRR72 pKa = 11.84GVSVKK77 pKa = 9.58TLSQHH82 pKa = 5.55ISQYY86 pKa = 9.8RR87 pKa = 11.84DD88 pKa = 2.95AYY90 pKa = 10.27QGEE93 pKa = 4.28LGLYY97 pKa = 8.48GVNSIVLCLALSGVNKK113 pKa = 10.27CQLTEE118 pKa = 3.72SSAYY122 pKa = 8.43HH123 pKa = 5.9FKK125 pKa = 9.73YY126 pKa = 9.04TSEE129 pKa = 3.85IATVCRR135 pKa = 11.84FFTQSDD141 pKa = 3.44KK142 pKa = 11.27TILFRR147 pKa = 11.84YY148 pKa = 8.49GRR150 pKa = 11.84VVSKK154 pKa = 10.32SAWSSLGIGHH164 pKa = 6.2KK165 pKa = 8.5TNWVFSVSLKK175 pKa = 7.64PTRR178 pKa = 11.84VSGLDD183 pKa = 3.7YY184 pKa = 10.82EE185 pKa = 4.47KK186 pKa = 10.64HH187 pKa = 5.49APKK190 pKa = 10.6AFAEE194 pKa = 4.35LFPLMEE200 pKa = 5.5LNFSRR205 pKa = 11.84DD206 pKa = 3.21PEE208 pKa = 4.39TNQIILVV215 pKa = 3.82
MM1 pKa = 7.34KK2 pKa = 10.42RR3 pKa = 11.84ILSKK7 pKa = 10.64SWWTSKK13 pKa = 9.9SHH15 pKa = 4.26SHH17 pKa = 6.07EE18 pKa = 4.19MGEE21 pKa = 4.5SSGSHH26 pKa = 6.97PEE28 pKa = 4.12SQSVPVSWDD37 pKa = 2.84QGVALTEE44 pKa = 4.35PPSAPLSSSGPFCLDD59 pKa = 3.58LEE61 pKa = 4.71VEE63 pKa = 4.72AYY65 pKa = 10.82LHH67 pKa = 5.23VQCNRR72 pKa = 11.84GVSVKK77 pKa = 9.58TLSQHH82 pKa = 5.55ISQYY86 pKa = 9.8RR87 pKa = 11.84DD88 pKa = 2.95AYY90 pKa = 10.27QGEE93 pKa = 4.28LGLYY97 pKa = 8.48GVNSIVLCLALSGVNKK113 pKa = 10.27CQLTEE118 pKa = 3.72SSAYY122 pKa = 8.43HH123 pKa = 5.9FKK125 pKa = 9.73YY126 pKa = 9.04TSEE129 pKa = 3.85IATVCRR135 pKa = 11.84FFTQSDD141 pKa = 3.44KK142 pKa = 11.27TILFRR147 pKa = 11.84YY148 pKa = 8.49GRR150 pKa = 11.84VVSKK154 pKa = 10.32SAWSSLGIGHH164 pKa = 6.2KK165 pKa = 8.5TNWVFSVSLKK175 pKa = 7.64PTRR178 pKa = 11.84VSGLDD183 pKa = 3.7YY184 pKa = 10.82EE185 pKa = 4.47KK186 pKa = 10.64HH187 pKa = 5.49APKK190 pKa = 10.6AFAEE194 pKa = 4.35LFPLMEE200 pKa = 5.5LNFSRR205 pKa = 11.84DD206 pKa = 3.21PEE208 pKa = 4.39TNQIILVV215 pKa = 3.82
Molecular weight: 23.94 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2904 |
215 |
2174 |
968.0 |
110.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.616 ± 0.418 | 1.86 ± 0.639 |
4.511 ± 0.488 | 6.267 ± 0.391 |
4.408 ± 0.352 | 5.82 ± 0.723 |
2.927 ± 0.345 | 7.025 ± 1.031 |
5.131 ± 0.249 | 10.572 ± 0.497 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.376 ± 0.357 | 4.787 ± 1.241 |
4.545 ± 0.261 | 3.099 ± 0.338 |
4.959 ± 0.593 | 9.539 ± 1.595 |
5.992 ± 0.326 | 6.198 ± 0.843 |
2.135 ± 0.054 | 4.236 ± 0.276 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
