
Candidatus Hodgkinia cicadicola
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Rhizobiales incertae sedis; Candidatus Hodgkinia
Average proteome isoelectric point is 8.34
Get precalculated fractions of proteins
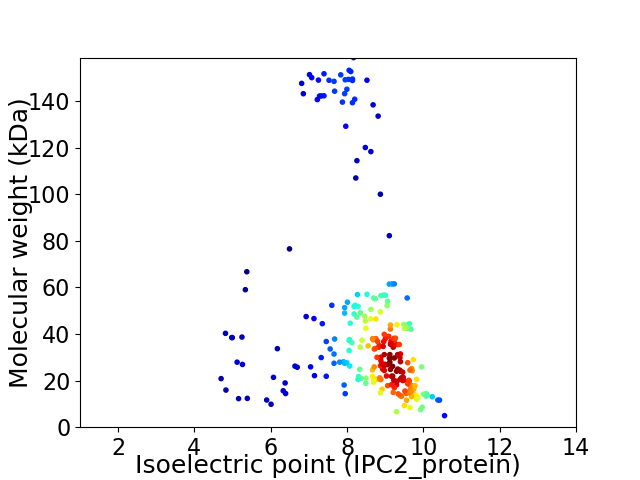
Virtual 2D-PAGE plot for 252 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2G9FSE3|A0A2G9FSE3_9RHIZ Phosphoribosyl-ATP diphosphatase OS=Candidatus Hodgkinia cicadicola OX=573658 GN=hisI PE=3 SV=1
MM1 pKa = 6.91FTGLVKK7 pKa = 9.95TLGMIVDD14 pKa = 4.22VLPLSGGIKK23 pKa = 10.18LCIISKK29 pKa = 10.13LRR31 pKa = 11.84LNVGNSISCSGVCLTVVSVNEE52 pKa = 4.0YY53 pKa = 9.02YY54 pKa = 10.81FEE56 pKa = 4.47VEE58 pKa = 3.96VWSEE62 pKa = 4.17SLCLSGLVDD71 pKa = 4.1LQRR74 pKa = 11.84FDD76 pKa = 3.56MVNLEE81 pKa = 4.25EE82 pKa = 5.74PITLNTPLHH91 pKa = 6.03GNITNGYY98 pKa = 7.85SKK100 pKa = 10.97SVVTIDD106 pKa = 2.99KK107 pKa = 9.62TYY109 pKa = 10.86VYY111 pKa = 10.02GDD113 pKa = 3.84SIILVIEE120 pKa = 4.2CPKK123 pKa = 9.87WISDD127 pKa = 4.14EE128 pKa = 4.07ITSPSSISLDD138 pKa = 3.41GVALTVTRR146 pKa = 11.84SGEE149 pKa = 4.03DD150 pKa = 3.36YY151 pKa = 11.32FEE153 pKa = 4.41VLLTRR158 pKa = 11.84YY159 pKa = 7.81TILNTTFKK167 pKa = 11.17YY168 pKa = 10.81FDD170 pKa = 3.4TKK172 pKa = 10.45RR173 pKa = 11.84WFSLEE178 pKa = 3.75QDD180 pKa = 3.67VSQRR184 pKa = 11.84WCC186 pKa = 3.47
MM1 pKa = 6.91FTGLVKK7 pKa = 9.95TLGMIVDD14 pKa = 4.22VLPLSGGIKK23 pKa = 10.18LCIISKK29 pKa = 10.13LRR31 pKa = 11.84LNVGNSISCSGVCLTVVSVNEE52 pKa = 4.0YY53 pKa = 9.02YY54 pKa = 10.81FEE56 pKa = 4.47VEE58 pKa = 3.96VWSEE62 pKa = 4.17SLCLSGLVDD71 pKa = 4.1LQRR74 pKa = 11.84FDD76 pKa = 3.56MVNLEE81 pKa = 4.25EE82 pKa = 5.74PITLNTPLHH91 pKa = 6.03GNITNGYY98 pKa = 7.85SKK100 pKa = 10.97SVVTIDD106 pKa = 2.99KK107 pKa = 9.62TYY109 pKa = 10.86VYY111 pKa = 10.02GDD113 pKa = 3.84SIILVIEE120 pKa = 4.2CPKK123 pKa = 9.87WISDD127 pKa = 4.14EE128 pKa = 4.07ITSPSSISLDD138 pKa = 3.41GVALTVTRR146 pKa = 11.84SGEE149 pKa = 4.03DD150 pKa = 3.36YY151 pKa = 11.32FEE153 pKa = 4.41VLLTRR158 pKa = 11.84YY159 pKa = 7.81TILNTTFKK167 pKa = 11.17YY168 pKa = 10.81FDD170 pKa = 3.4TKK172 pKa = 10.45RR173 pKa = 11.84WFSLEE178 pKa = 3.75QDD180 pKa = 3.67VSQRR184 pKa = 11.84WCC186 pKa = 3.47
Molecular weight: 20.85 kDa
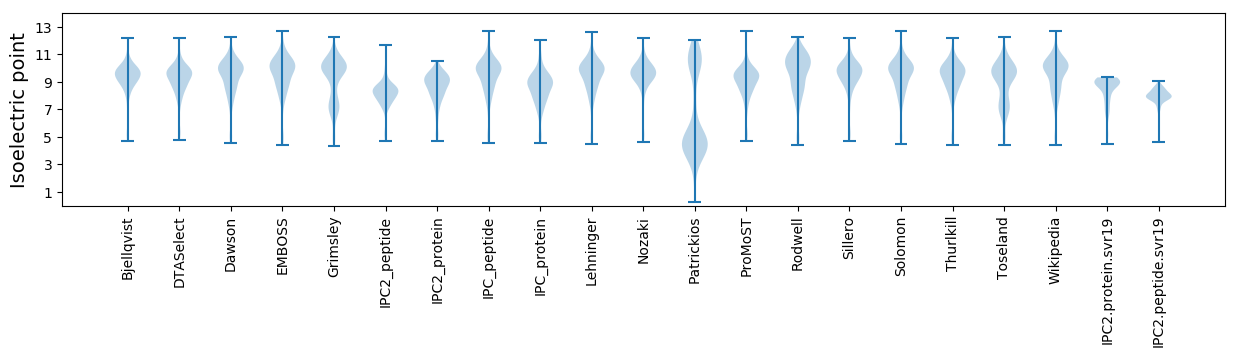
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2G9FSF5|A0A2G9FSF5_9RHIZ Electron transfer flavoprotein large subunit OS=Candidatus Hodgkinia cicadicola OX=573658 GN=etfA PE=4 SV=1
MM1 pKa = 8.07RR2 pKa = 11.84YY3 pKa = 9.25NVQINVRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84PWRR15 pKa = 11.84SWSCFGLYY23 pKa = 10.21RR24 pKa = 11.84LIDD27 pKa = 3.71LVSIKK32 pKa = 10.53SQILSDD38 pKa = 3.63RR39 pKa = 11.84NSWITEE45 pKa = 4.09LLTTRR50 pKa = 11.84PWFIFKK56 pKa = 10.4RR57 pKa = 11.84IKK59 pKa = 9.84EE60 pKa = 4.16EE61 pKa = 4.21QMEE64 pKa = 4.23LTLAVNFQVNRR75 pKa = 11.84KK76 pKa = 8.75IVIEE80 pKa = 4.5SIDD83 pKa = 4.66LILQVFLLARR93 pKa = 11.84RR94 pKa = 11.84IGLNINNVIRR104 pKa = 11.84LIDD107 pKa = 3.45YY108 pKa = 8.41WNIVASTQRR117 pKa = 11.84FNKK120 pKa = 8.46WSNFSRR126 pKa = 11.84PNKK129 pKa = 10.21SNYY132 pKa = 5.87NTNYY136 pKa = 9.35RR137 pKa = 11.84SSMNNFRR144 pKa = 11.84SNRR147 pKa = 11.84LDD149 pKa = 4.03NITLSRR155 pKa = 11.84LNNSISNLFMVITNLRR171 pKa = 11.84TGSYY175 pKa = 9.86RR176 pKa = 11.84YY177 pKa = 10.18RR178 pKa = 11.84NILNGLFYY186 pKa = 11.12NVLYY190 pKa = 10.96NIITLICNRR199 pKa = 11.84SIKK202 pKa = 10.45YY203 pKa = 10.74SMVINEE209 pKa = 5.35IIDD212 pKa = 3.42KK213 pKa = 10.38MEE215 pKa = 3.79
MM1 pKa = 8.07RR2 pKa = 11.84YY3 pKa = 9.25NVQINVRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84PWRR15 pKa = 11.84SWSCFGLYY23 pKa = 10.21RR24 pKa = 11.84LIDD27 pKa = 3.71LVSIKK32 pKa = 10.53SQILSDD38 pKa = 3.63RR39 pKa = 11.84NSWITEE45 pKa = 4.09LLTTRR50 pKa = 11.84PWFIFKK56 pKa = 10.4RR57 pKa = 11.84IKK59 pKa = 9.84EE60 pKa = 4.16EE61 pKa = 4.21QMEE64 pKa = 4.23LTLAVNFQVNRR75 pKa = 11.84KK76 pKa = 8.75IVIEE80 pKa = 4.5SIDD83 pKa = 4.66LILQVFLLARR93 pKa = 11.84RR94 pKa = 11.84IGLNINNVIRR104 pKa = 11.84LIDD107 pKa = 3.45YY108 pKa = 8.41WNIVASTQRR117 pKa = 11.84FNKK120 pKa = 8.46WSNFSRR126 pKa = 11.84PNKK129 pKa = 10.21SNYY132 pKa = 5.87NTNYY136 pKa = 9.35RR137 pKa = 11.84SSMNNFRR144 pKa = 11.84SNRR147 pKa = 11.84LDD149 pKa = 4.03NITLSRR155 pKa = 11.84LNNSISNLFMVITNLRR171 pKa = 11.84TGSYY175 pKa = 9.86RR176 pKa = 11.84YY177 pKa = 10.18RR178 pKa = 11.84NILNGLFYY186 pKa = 11.12NVLYY190 pKa = 10.96NIITLICNRR199 pKa = 11.84SIKK202 pKa = 10.45YY203 pKa = 10.74SMVINEE209 pKa = 5.35IIDD212 pKa = 3.42KK213 pKa = 10.38MEE215 pKa = 3.79
Molecular weight: 25.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
101011 |
41 |
1395 |
400.8 |
45.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.181 ± 0.107 | 2.308 ± 0.056 |
4.682 ± 0.126 | 4.196 ± 0.097 |
3.778 ± 0.094 | 5.518 ± 0.133 |
2.057 ± 0.044 | 10.031 ± 0.164 |
7.942 ± 0.127 | 10.854 ± 0.111 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.427 ± 0.051 | 7.646 ± 0.119 |
3.063 ± 0.058 | 2.715 ± 0.065 |
4.541 ± 0.089 | 7.608 ± 0.106 |
5.958 ± 0.115 | 6.611 ± 0.146 |
1.215 ± 0.046 | 3.671 ± 0.079 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
