
Beihai weivirus-like virus 5
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.19
Get precalculated fractions of proteins
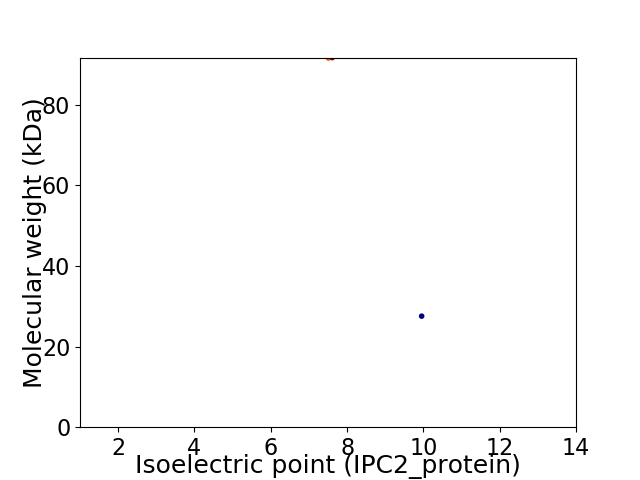
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KL21|A0A1L3KL21_9VIRU RNA-dependent RNA polymerase OS=Beihai weivirus-like virus 5 OX=1922753 PE=4 SV=1
MM1 pKa = 7.62LPLRR5 pKa = 11.84SGNCDD10 pKa = 2.82LTLRR14 pKa = 11.84DD15 pKa = 3.77VPHH18 pKa = 6.44YY19 pKa = 10.49LQAQAGTRR27 pKa = 11.84KK28 pKa = 9.57GAWEE32 pKa = 3.81ITVYY36 pKa = 10.58EE37 pKa = 4.88HH38 pKa = 6.81DD39 pKa = 4.43AEE41 pKa = 4.53WFKK44 pKa = 11.23SWRR47 pKa = 11.84PNAYY51 pKa = 9.18SVRR54 pKa = 11.84FTEE57 pKa = 5.17DD58 pKa = 3.07EE59 pKa = 4.06YY60 pKa = 11.89SKK62 pKa = 11.07LSRR65 pKa = 11.84TMYY68 pKa = 10.01HH69 pKa = 6.74GLRR72 pKa = 11.84NGTKK76 pKa = 9.03LTSLYY81 pKa = 10.87AGLEE85 pKa = 4.12NASQSLWPTGSLDD98 pKa = 3.59HH99 pKa = 6.72VAMRR103 pKa = 11.84HH104 pKa = 4.78MGFPTVWRR112 pKa = 11.84YY113 pKa = 7.62ITNQTPARR121 pKa = 11.84FEE123 pKa = 4.11FAHH126 pKa = 6.95KK127 pKa = 10.55LGTYY131 pKa = 10.11LVAPQSLLVPWWMITKK147 pKa = 10.33KK148 pKa = 8.94MRR150 pKa = 11.84RR151 pKa = 11.84RR152 pKa = 11.84YY153 pKa = 9.46AVAAFLVLAGLALKK167 pKa = 10.29LRR169 pKa = 11.84RR170 pKa = 11.84IGRR173 pKa = 11.84LALPRR178 pKa = 11.84LTRR181 pKa = 11.84VTGQSGGQEE190 pKa = 3.91AKK192 pKa = 10.34KK193 pKa = 10.41DD194 pKa = 3.79DD195 pKa = 4.28PSGGKK200 pKa = 9.37PPTPNEE206 pKa = 4.3GGPKK210 pKa = 10.5DD211 pKa = 4.47GDD213 pKa = 3.82TTPDD217 pKa = 3.2AGNVKK222 pKa = 10.42PEE224 pKa = 4.39GGNNDD229 pKa = 4.62DD230 pKa = 4.61PPAPPMTTLRR240 pKa = 11.84RR241 pKa = 11.84TDD243 pKa = 3.03TSFVIGGPPVMDD255 pKa = 4.55EE256 pKa = 4.05PALPGDD262 pKa = 4.25FEE264 pKa = 4.48NAAEE268 pKa = 4.02AQEE271 pKa = 4.43DD272 pKa = 3.91GRR274 pKa = 11.84TVVRR278 pKa = 11.84GDD280 pKa = 3.33VVAVLGQYY288 pKa = 9.86FDD290 pKa = 3.62KK291 pKa = 10.87DD292 pKa = 3.31RR293 pKa = 11.84PINHH297 pKa = 5.95MPIVGCMVGPCQLKK311 pKa = 9.35PNVYY315 pKa = 10.56AKK317 pKa = 9.73TVFNLWAAIVKK328 pKa = 10.05RR329 pKa = 11.84IRR331 pKa = 11.84EE332 pKa = 4.0KK333 pKa = 10.82ALKK336 pKa = 9.34PKK338 pKa = 10.74LSGEE342 pKa = 3.95EE343 pKa = 3.64KK344 pKa = 9.98TRR346 pKa = 11.84IGRR349 pKa = 11.84LISKK353 pKa = 10.19CMTPDD358 pKa = 2.94KK359 pKa = 10.63RR360 pKa = 11.84CGVFAKK366 pKa = 10.39HH367 pKa = 6.69RR368 pKa = 11.84IEE370 pKa = 4.1EE371 pKa = 3.95WAIAHH376 pKa = 6.94FDD378 pKa = 4.02LEE380 pKa = 4.5EE381 pKa = 4.4CKK383 pKa = 10.52SGKK386 pKa = 9.21WSIEE390 pKa = 3.74RR391 pKa = 11.84FRR393 pKa = 11.84GSLEE397 pKa = 3.48NLYY400 pKa = 10.87AKK402 pKa = 8.4EE403 pKa = 4.19HH404 pKa = 5.94PTFSFKK410 pKa = 11.16ADD412 pKa = 3.24VKK414 pKa = 9.61YY415 pKa = 11.13EE416 pKa = 3.95CMPEE420 pKa = 4.02GKK422 pKa = 10.12APRR425 pKa = 11.84MLIADD430 pKa = 3.91GDD432 pKa = 4.11EE433 pKa = 4.41GQLMALAVVKK443 pKa = 10.57CFEE446 pKa = 4.38EE447 pKa = 4.42LLFSHH452 pKa = 7.42FEE454 pKa = 4.18TKK456 pKa = 10.23SIKK459 pKa = 10.31HH460 pKa = 5.14LAKK463 pKa = 9.81RR464 pKa = 11.84DD465 pKa = 3.9AIDD468 pKa = 3.58RR469 pKa = 11.84VLKK472 pKa = 9.98EE473 pKa = 4.0LRR475 pKa = 11.84APGAKK480 pKa = 9.58AVEE483 pKa = 4.6GDD485 pKa = 3.29GSAWDD490 pKa = 3.89TTCNVLIRR498 pKa = 11.84GLVEE502 pKa = 4.04NPVLRR507 pKa = 11.84HH508 pKa = 5.04ITTVLCNFGVIPSTWMEE525 pKa = 3.72EE526 pKa = 3.91HH527 pKa = 6.49QRR529 pKa = 11.84ACEE532 pKa = 3.94KK533 pKa = 9.79KK534 pKa = 7.57TLRR537 pKa = 11.84LFFSNKK543 pKa = 9.0FEE545 pKa = 4.26TMSTSIDD552 pKa = 3.56AIRR555 pKa = 11.84RR556 pKa = 11.84SGHH559 pKa = 6.55RR560 pKa = 11.84GTSCLNWWINFVLWVSSVFKK580 pKa = 10.52EE581 pKa = 4.05PEE583 pKa = 3.76RR584 pKa = 11.84FLDD587 pKa = 3.49VAVRR591 pKa = 11.84NGTDD595 pKa = 3.05LTGRR599 pKa = 11.84SRR601 pKa = 11.84WWNGCFEE608 pKa = 5.17GDD610 pKa = 5.07DD611 pKa = 4.13SLCTMRR617 pKa = 11.84PPMVEE622 pKa = 4.14GDD624 pKa = 3.87ALCQVFLAFWKK635 pKa = 10.68SAGFNMKK642 pKa = 9.66IVFCKK647 pKa = 9.9TRR649 pKa = 11.84ATFVGWHH656 pKa = 5.7VGCTDD661 pKa = 3.47GEE663 pKa = 4.59LNNFRR668 pKa = 11.84CPEE671 pKa = 4.08LPRR674 pKa = 11.84ALANSGVSVSPEE686 pKa = 3.89AIKK689 pKa = 10.38AAKK692 pKa = 10.11DD693 pKa = 3.51MNRR696 pKa = 11.84SAVNVLAAASALARR710 pKa = 11.84ASDD713 pKa = 3.79FAGILPSVSVKK724 pKa = 10.91YY725 pKa = 10.01MDD727 pKa = 3.77FAEE730 pKa = 4.59SVSRR734 pKa = 11.84TDD736 pKa = 3.68FSDD739 pKa = 3.82RR740 pKa = 11.84EE741 pKa = 3.78MSIRR745 pKa = 11.84AFGEE749 pKa = 4.07DD750 pKa = 3.51GFSANAVRR758 pKa = 11.84TQIMEE763 pKa = 4.17RR764 pKa = 11.84NIGITPDD771 pKa = 3.47EE772 pKa = 4.5EE773 pKa = 4.77LVTMAALGYY782 pKa = 10.41AATQDD787 pKa = 3.44EE788 pKa = 5.21VATFMEE794 pKa = 5.13YY795 pKa = 10.52GWSLDD800 pKa = 3.34PHH802 pKa = 6.74VLLDD806 pKa = 3.58YY807 pKa = 10.78EE808 pKa = 4.49AFRR811 pKa = 11.84GSLPPSWRR819 pKa = 11.84AA820 pKa = 2.98
MM1 pKa = 7.62LPLRR5 pKa = 11.84SGNCDD10 pKa = 2.82LTLRR14 pKa = 11.84DD15 pKa = 3.77VPHH18 pKa = 6.44YY19 pKa = 10.49LQAQAGTRR27 pKa = 11.84KK28 pKa = 9.57GAWEE32 pKa = 3.81ITVYY36 pKa = 10.58EE37 pKa = 4.88HH38 pKa = 6.81DD39 pKa = 4.43AEE41 pKa = 4.53WFKK44 pKa = 11.23SWRR47 pKa = 11.84PNAYY51 pKa = 9.18SVRR54 pKa = 11.84FTEE57 pKa = 5.17DD58 pKa = 3.07EE59 pKa = 4.06YY60 pKa = 11.89SKK62 pKa = 11.07LSRR65 pKa = 11.84TMYY68 pKa = 10.01HH69 pKa = 6.74GLRR72 pKa = 11.84NGTKK76 pKa = 9.03LTSLYY81 pKa = 10.87AGLEE85 pKa = 4.12NASQSLWPTGSLDD98 pKa = 3.59HH99 pKa = 6.72VAMRR103 pKa = 11.84HH104 pKa = 4.78MGFPTVWRR112 pKa = 11.84YY113 pKa = 7.62ITNQTPARR121 pKa = 11.84FEE123 pKa = 4.11FAHH126 pKa = 6.95KK127 pKa = 10.55LGTYY131 pKa = 10.11LVAPQSLLVPWWMITKK147 pKa = 10.33KK148 pKa = 8.94MRR150 pKa = 11.84RR151 pKa = 11.84RR152 pKa = 11.84YY153 pKa = 9.46AVAAFLVLAGLALKK167 pKa = 10.29LRR169 pKa = 11.84RR170 pKa = 11.84IGRR173 pKa = 11.84LALPRR178 pKa = 11.84LTRR181 pKa = 11.84VTGQSGGQEE190 pKa = 3.91AKK192 pKa = 10.34KK193 pKa = 10.41DD194 pKa = 3.79DD195 pKa = 4.28PSGGKK200 pKa = 9.37PPTPNEE206 pKa = 4.3GGPKK210 pKa = 10.5DD211 pKa = 4.47GDD213 pKa = 3.82TTPDD217 pKa = 3.2AGNVKK222 pKa = 10.42PEE224 pKa = 4.39GGNNDD229 pKa = 4.62DD230 pKa = 4.61PPAPPMTTLRR240 pKa = 11.84RR241 pKa = 11.84TDD243 pKa = 3.03TSFVIGGPPVMDD255 pKa = 4.55EE256 pKa = 4.05PALPGDD262 pKa = 4.25FEE264 pKa = 4.48NAAEE268 pKa = 4.02AQEE271 pKa = 4.43DD272 pKa = 3.91GRR274 pKa = 11.84TVVRR278 pKa = 11.84GDD280 pKa = 3.33VVAVLGQYY288 pKa = 9.86FDD290 pKa = 3.62KK291 pKa = 10.87DD292 pKa = 3.31RR293 pKa = 11.84PINHH297 pKa = 5.95MPIVGCMVGPCQLKK311 pKa = 9.35PNVYY315 pKa = 10.56AKK317 pKa = 9.73TVFNLWAAIVKK328 pKa = 10.05RR329 pKa = 11.84IRR331 pKa = 11.84EE332 pKa = 4.0KK333 pKa = 10.82ALKK336 pKa = 9.34PKK338 pKa = 10.74LSGEE342 pKa = 3.95EE343 pKa = 3.64KK344 pKa = 9.98TRR346 pKa = 11.84IGRR349 pKa = 11.84LISKK353 pKa = 10.19CMTPDD358 pKa = 2.94KK359 pKa = 10.63RR360 pKa = 11.84CGVFAKK366 pKa = 10.39HH367 pKa = 6.69RR368 pKa = 11.84IEE370 pKa = 4.1EE371 pKa = 3.95WAIAHH376 pKa = 6.94FDD378 pKa = 4.02LEE380 pKa = 4.5EE381 pKa = 4.4CKK383 pKa = 10.52SGKK386 pKa = 9.21WSIEE390 pKa = 3.74RR391 pKa = 11.84FRR393 pKa = 11.84GSLEE397 pKa = 3.48NLYY400 pKa = 10.87AKK402 pKa = 8.4EE403 pKa = 4.19HH404 pKa = 5.94PTFSFKK410 pKa = 11.16ADD412 pKa = 3.24VKK414 pKa = 9.61YY415 pKa = 11.13EE416 pKa = 3.95CMPEE420 pKa = 4.02GKK422 pKa = 10.12APRR425 pKa = 11.84MLIADD430 pKa = 3.91GDD432 pKa = 4.11EE433 pKa = 4.41GQLMALAVVKK443 pKa = 10.57CFEE446 pKa = 4.38EE447 pKa = 4.42LLFSHH452 pKa = 7.42FEE454 pKa = 4.18TKK456 pKa = 10.23SIKK459 pKa = 10.31HH460 pKa = 5.14LAKK463 pKa = 9.81RR464 pKa = 11.84DD465 pKa = 3.9AIDD468 pKa = 3.58RR469 pKa = 11.84VLKK472 pKa = 9.98EE473 pKa = 4.0LRR475 pKa = 11.84APGAKK480 pKa = 9.58AVEE483 pKa = 4.6GDD485 pKa = 3.29GSAWDD490 pKa = 3.89TTCNVLIRR498 pKa = 11.84GLVEE502 pKa = 4.04NPVLRR507 pKa = 11.84HH508 pKa = 5.04ITTVLCNFGVIPSTWMEE525 pKa = 3.72EE526 pKa = 3.91HH527 pKa = 6.49QRR529 pKa = 11.84ACEE532 pKa = 3.94KK533 pKa = 9.79KK534 pKa = 7.57TLRR537 pKa = 11.84LFFSNKK543 pKa = 9.0FEE545 pKa = 4.26TMSTSIDD552 pKa = 3.56AIRR555 pKa = 11.84RR556 pKa = 11.84SGHH559 pKa = 6.55RR560 pKa = 11.84GTSCLNWWINFVLWVSSVFKK580 pKa = 10.52EE581 pKa = 4.05PEE583 pKa = 3.76RR584 pKa = 11.84FLDD587 pKa = 3.49VAVRR591 pKa = 11.84NGTDD595 pKa = 3.05LTGRR599 pKa = 11.84SRR601 pKa = 11.84WWNGCFEE608 pKa = 5.17GDD610 pKa = 5.07DD611 pKa = 4.13SLCTMRR617 pKa = 11.84PPMVEE622 pKa = 4.14GDD624 pKa = 3.87ALCQVFLAFWKK635 pKa = 10.68SAGFNMKK642 pKa = 9.66IVFCKK647 pKa = 9.9TRR649 pKa = 11.84ATFVGWHH656 pKa = 5.7VGCTDD661 pKa = 3.47GEE663 pKa = 4.59LNNFRR668 pKa = 11.84CPEE671 pKa = 4.08LPRR674 pKa = 11.84ALANSGVSVSPEE686 pKa = 3.89AIKK689 pKa = 10.38AAKK692 pKa = 10.11DD693 pKa = 3.51MNRR696 pKa = 11.84SAVNVLAAASALARR710 pKa = 11.84ASDD713 pKa = 3.79FAGILPSVSVKK724 pKa = 10.91YY725 pKa = 10.01MDD727 pKa = 3.77FAEE730 pKa = 4.59SVSRR734 pKa = 11.84TDD736 pKa = 3.68FSDD739 pKa = 3.82RR740 pKa = 11.84EE741 pKa = 3.78MSIRR745 pKa = 11.84AFGEE749 pKa = 4.07DD750 pKa = 3.51GFSANAVRR758 pKa = 11.84TQIMEE763 pKa = 4.17RR764 pKa = 11.84NIGITPDD771 pKa = 3.47EE772 pKa = 4.5EE773 pKa = 4.77LVTMAALGYY782 pKa = 10.41AATQDD787 pKa = 3.44EE788 pKa = 5.21VATFMEE794 pKa = 5.13YY795 pKa = 10.52GWSLDD800 pKa = 3.34PHH802 pKa = 6.74VLLDD806 pKa = 3.58YY807 pKa = 10.78EE808 pKa = 4.49AFRR811 pKa = 11.84GSLPPSWRR819 pKa = 11.84AA820 pKa = 2.98
Molecular weight: 91.58 kDa
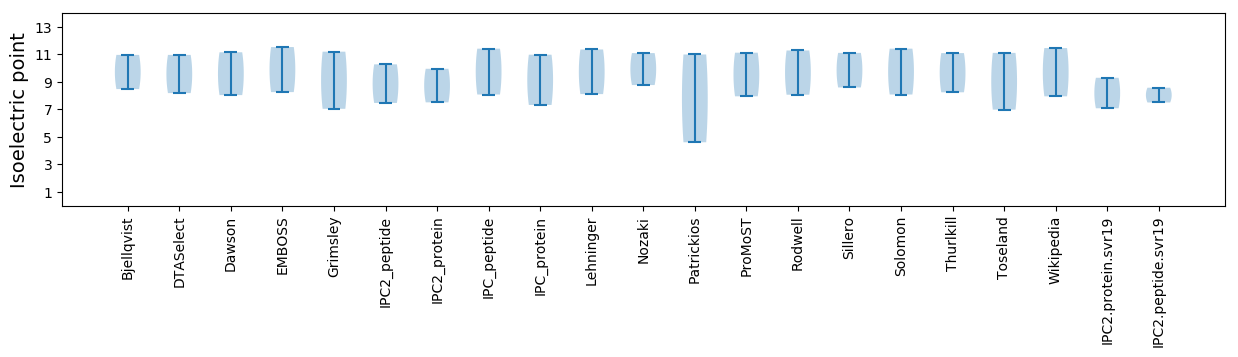
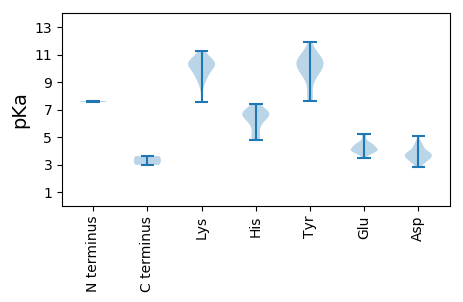
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KL21|A0A1L3KL21_9VIRU RNA-dependent RNA polymerase OS=Beihai weivirus-like virus 5 OX=1922753 PE=4 SV=1
MM1 pKa = 7.55AKK3 pKa = 9.94GRR5 pKa = 11.84SSLKK9 pKa = 10.02PVTQARR15 pKa = 11.84RR16 pKa = 11.84WNRR19 pKa = 11.84RR20 pKa = 11.84KK21 pKa = 10.55GGGVTRR27 pKa = 11.84ATAPRR32 pKa = 11.84VLAQGACAVPKK43 pKa = 10.3KK44 pKa = 10.92AFGAGKK50 pKa = 9.95NPNGWKK56 pKa = 10.21KK57 pKa = 11.2AMTLGLNARR66 pKa = 11.84LPYY69 pKa = 10.31HH70 pKa = 6.99LGLPRR75 pKa = 11.84PVGPYY80 pKa = 9.52QVIRR84 pKa = 11.84TTKK87 pKa = 9.29VHH89 pKa = 4.94TTKK92 pKa = 10.42SGCVIFTPLQRR103 pKa = 11.84RR104 pKa = 11.84HH105 pKa = 6.63DD106 pKa = 4.24SLPGEE111 pKa = 4.29PRR113 pKa = 11.84WYY115 pKa = 9.88AACGMEE121 pKa = 4.27AVNTGVSINAANNATMISMPMDD143 pKa = 3.68VLGSACDD150 pKa = 3.67VVPAAMSVQVMCPTALQNASGAFTMARR177 pKa = 11.84VNQQLTFGGDD187 pKa = 3.29TRR189 pKa = 11.84PWGQFITQFNSFFRR203 pKa = 11.84PRR205 pKa = 11.84LLTGGKK211 pKa = 8.68LALRR215 pKa = 11.84GVHH218 pKa = 6.33ANAYY222 pKa = 7.79PLDD225 pKa = 3.76MSEE228 pKa = 4.24YY229 pKa = 8.35AHH231 pKa = 6.52FAGVVPSLGTTTWNAITQSATRR253 pKa = 11.84LPAVV257 pKa = 3.62
MM1 pKa = 7.55AKK3 pKa = 9.94GRR5 pKa = 11.84SSLKK9 pKa = 10.02PVTQARR15 pKa = 11.84RR16 pKa = 11.84WNRR19 pKa = 11.84RR20 pKa = 11.84KK21 pKa = 10.55GGGVTRR27 pKa = 11.84ATAPRR32 pKa = 11.84VLAQGACAVPKK43 pKa = 10.3KK44 pKa = 10.92AFGAGKK50 pKa = 9.95NPNGWKK56 pKa = 10.21KK57 pKa = 11.2AMTLGLNARR66 pKa = 11.84LPYY69 pKa = 10.31HH70 pKa = 6.99LGLPRR75 pKa = 11.84PVGPYY80 pKa = 9.52QVIRR84 pKa = 11.84TTKK87 pKa = 9.29VHH89 pKa = 4.94TTKK92 pKa = 10.42SGCVIFTPLQRR103 pKa = 11.84RR104 pKa = 11.84HH105 pKa = 6.63DD106 pKa = 4.24SLPGEE111 pKa = 4.29PRR113 pKa = 11.84WYY115 pKa = 9.88AACGMEE121 pKa = 4.27AVNTGVSINAANNATMISMPMDD143 pKa = 3.68VLGSACDD150 pKa = 3.67VVPAAMSVQVMCPTALQNASGAFTMARR177 pKa = 11.84VNQQLTFGGDD187 pKa = 3.29TRR189 pKa = 11.84PWGQFITQFNSFFRR203 pKa = 11.84PRR205 pKa = 11.84LLTGGKK211 pKa = 8.68LALRR215 pKa = 11.84GVHH218 pKa = 6.33ANAYY222 pKa = 7.79PLDD225 pKa = 3.76MSEE228 pKa = 4.24YY229 pKa = 8.35AHH231 pKa = 6.52FAGVVPSLGTTTWNAITQSATRR253 pKa = 11.84LPAVV257 pKa = 3.62
Molecular weight: 27.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1077 |
257 |
820 |
538.5 |
59.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.935 ± 1.277 | 2.136 ± 0.096 |
4.643 ± 1.369 | 5.2 ± 2.046 |
4.457 ± 0.485 | 8.078 ± 0.837 |
2.043 ± 0.049 | 3.343 ± 0.512 |
5.292 ± 0.514 | 8.078 ± 0.348 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.25 ± 0.325 | 3.993 ± 0.541 |
6.128 ± 0.642 | 2.414 ± 0.947 |
7.242 ± 0.076 | 5.757 ± 0.354 |
6.592 ± 0.999 | 6.964 ± 0.415 |
2.414 ± 0.238 | 2.043 ± 0.049 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
