
Daeseongdong virus 1
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 6.93
Get precalculated fractions of proteins
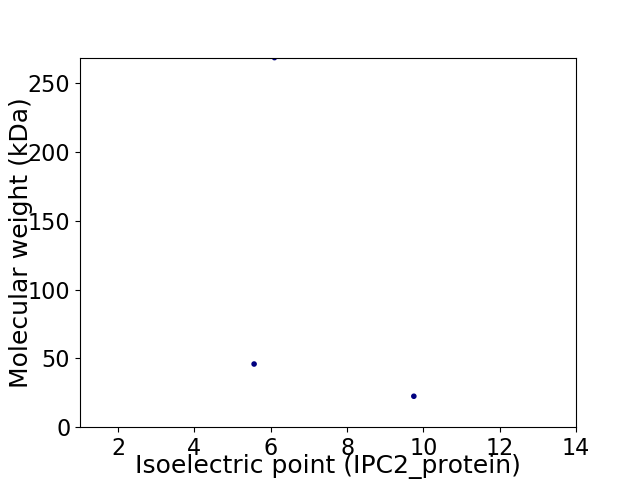
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
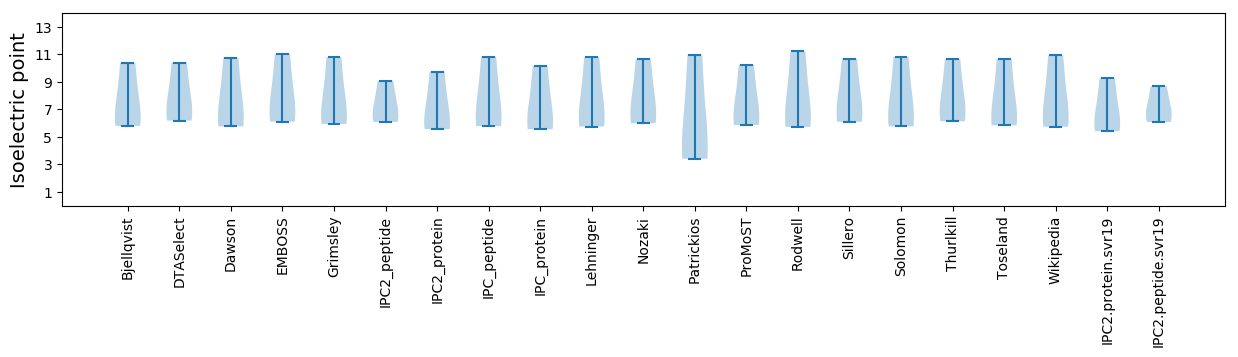
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0S2RRE2|A0A0S2RRE2_9VIRU DiSB-ORF2_chro domain-containing protein OS=Daeseongdong virus 1 OX=1758881 PE=4 SV=1
MM1 pKa = 7.69KK2 pKa = 10.59SFLLFVLFLTTSVHH16 pKa = 5.58AAVFIKK22 pKa = 10.67DD23 pKa = 3.28IVRR26 pKa = 11.84QLTATRR32 pKa = 11.84EE33 pKa = 3.84KK34 pKa = 10.62HH35 pKa = 5.96LLYY38 pKa = 10.86SKK40 pKa = 10.82LQMRR44 pKa = 11.84LHH46 pKa = 6.32FVGEE50 pKa = 4.31FDD52 pKa = 3.05ISPYY56 pKa = 10.82KK57 pKa = 10.2FDD59 pKa = 4.33PEE61 pKa = 4.28CLSVYY66 pKa = 8.85RR67 pKa = 11.84TNDD70 pKa = 2.56WFFSKK75 pKa = 11.0CEE77 pKa = 4.01LPSNCSKK84 pKa = 10.69PLVDD88 pKa = 4.86VIDD91 pKa = 4.5RR92 pKa = 11.84NWLGQEE98 pKa = 3.97TVLCHH103 pKa = 5.85GVHH106 pKa = 6.84PSEE109 pKa = 4.47TSDD112 pKa = 2.96RR113 pKa = 11.84MNFFSMEE120 pKa = 4.1FVPSNFTFGRR130 pKa = 11.84PITIDD135 pKa = 2.84GRR137 pKa = 11.84EE138 pKa = 3.94FTRR141 pKa = 11.84FEE143 pKa = 4.4DD144 pKa = 4.38FVPVSFFEE152 pKa = 3.8WFYY155 pKa = 10.39MVKK158 pKa = 10.38SPGDD162 pKa = 3.4SYY164 pKa = 11.85GIVPKK169 pKa = 10.43FCSEE173 pKa = 4.35TYY175 pKa = 10.12RR176 pKa = 11.84DD177 pKa = 4.11STTLPPNVNVKK188 pKa = 10.45SDD190 pKa = 3.47GNTLCLTEE198 pKa = 3.93IQYY201 pKa = 10.4DD202 pKa = 3.81QSDD205 pKa = 4.86CRR207 pKa = 11.84PKK209 pKa = 10.08WYY211 pKa = 9.25PSYY214 pKa = 11.01HH215 pKa = 6.83SIFADD220 pKa = 2.6IDD222 pKa = 4.01FPVEE226 pKa = 4.02FSGVPYY232 pKa = 10.86SPGSYY237 pKa = 9.8SATNNKK243 pKa = 9.41VIGADD248 pKa = 3.2VYY250 pKa = 11.21MKK252 pKa = 10.81DD253 pKa = 3.52NLGNTPTHH261 pKa = 6.51VIDD264 pKa = 4.17KK265 pKa = 10.93VGGAYY270 pKa = 8.72MVTYY274 pKa = 10.37SPYY277 pKa = 10.37NGLGPIYY284 pKa = 9.68IVKK287 pKa = 8.59ATSVIPKK294 pKa = 9.74GDD296 pKa = 3.48SRR298 pKa = 11.84CFEE301 pKa = 4.11LQSRR305 pKa = 11.84YY306 pKa = 9.69HH307 pKa = 7.19SPFEE311 pKa = 3.63ILIAKK316 pKa = 9.47ISKK319 pKa = 9.67FFRR322 pKa = 11.84QEE324 pKa = 3.48LEE326 pKa = 3.94YY327 pKa = 11.21LLEE330 pKa = 3.9FLIAFAEE337 pKa = 4.71KK338 pKa = 9.99IASILIAILGEE349 pKa = 3.78IMNVVLSLVPYY360 pKa = 10.6GDD362 pKa = 4.29LFYY365 pKa = 10.91TSAFLAAVTYY375 pKa = 10.53LFTRR379 pKa = 11.84DD380 pKa = 3.35VILALVPNIAIYY392 pKa = 10.03LVRR395 pKa = 11.84IYY397 pKa = 10.62LKK399 pKa = 10.81SAII402 pKa = 4.18
MM1 pKa = 7.69KK2 pKa = 10.59SFLLFVLFLTTSVHH16 pKa = 5.58AAVFIKK22 pKa = 10.67DD23 pKa = 3.28IVRR26 pKa = 11.84QLTATRR32 pKa = 11.84EE33 pKa = 3.84KK34 pKa = 10.62HH35 pKa = 5.96LLYY38 pKa = 10.86SKK40 pKa = 10.82LQMRR44 pKa = 11.84LHH46 pKa = 6.32FVGEE50 pKa = 4.31FDD52 pKa = 3.05ISPYY56 pKa = 10.82KK57 pKa = 10.2FDD59 pKa = 4.33PEE61 pKa = 4.28CLSVYY66 pKa = 8.85RR67 pKa = 11.84TNDD70 pKa = 2.56WFFSKK75 pKa = 11.0CEE77 pKa = 4.01LPSNCSKK84 pKa = 10.69PLVDD88 pKa = 4.86VIDD91 pKa = 4.5RR92 pKa = 11.84NWLGQEE98 pKa = 3.97TVLCHH103 pKa = 5.85GVHH106 pKa = 6.84PSEE109 pKa = 4.47TSDD112 pKa = 2.96RR113 pKa = 11.84MNFFSMEE120 pKa = 4.1FVPSNFTFGRR130 pKa = 11.84PITIDD135 pKa = 2.84GRR137 pKa = 11.84EE138 pKa = 3.94FTRR141 pKa = 11.84FEE143 pKa = 4.4DD144 pKa = 4.38FVPVSFFEE152 pKa = 3.8WFYY155 pKa = 10.39MVKK158 pKa = 10.38SPGDD162 pKa = 3.4SYY164 pKa = 11.85GIVPKK169 pKa = 10.43FCSEE173 pKa = 4.35TYY175 pKa = 10.12RR176 pKa = 11.84DD177 pKa = 4.11STTLPPNVNVKK188 pKa = 10.45SDD190 pKa = 3.47GNTLCLTEE198 pKa = 3.93IQYY201 pKa = 10.4DD202 pKa = 3.81QSDD205 pKa = 4.86CRR207 pKa = 11.84PKK209 pKa = 10.08WYY211 pKa = 9.25PSYY214 pKa = 11.01HH215 pKa = 6.83SIFADD220 pKa = 2.6IDD222 pKa = 4.01FPVEE226 pKa = 4.02FSGVPYY232 pKa = 10.86SPGSYY237 pKa = 9.8SATNNKK243 pKa = 9.41VIGADD248 pKa = 3.2VYY250 pKa = 11.21MKK252 pKa = 10.81DD253 pKa = 3.52NLGNTPTHH261 pKa = 6.51VIDD264 pKa = 4.17KK265 pKa = 10.93VGGAYY270 pKa = 8.72MVTYY274 pKa = 10.37SPYY277 pKa = 10.37NGLGPIYY284 pKa = 9.68IVKK287 pKa = 8.59ATSVIPKK294 pKa = 9.74GDD296 pKa = 3.48SRR298 pKa = 11.84CFEE301 pKa = 4.11LQSRR305 pKa = 11.84YY306 pKa = 9.69HH307 pKa = 7.19SPFEE311 pKa = 3.63ILIAKK316 pKa = 9.47ISKK319 pKa = 9.67FFRR322 pKa = 11.84QEE324 pKa = 3.48LEE326 pKa = 3.94YY327 pKa = 11.21LLEE330 pKa = 3.9FLIAFAEE337 pKa = 4.71KK338 pKa = 9.99IASILIAILGEE349 pKa = 3.78IMNVVLSLVPYY360 pKa = 10.6GDD362 pKa = 4.29LFYY365 pKa = 10.91TSAFLAAVTYY375 pKa = 10.53LFTRR379 pKa = 11.84DD380 pKa = 3.35VILALVPNIAIYY392 pKa = 10.03LVRR395 pKa = 11.84IYY397 pKa = 10.62LKK399 pKa = 10.81SAII402 pKa = 4.18
Molecular weight: 45.89 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0S2RRD3|A0A0S2RRD3_9VIRU Non-structural protein 3 OS=Daeseongdong virus 1 OX=1758881 PE=4 SV=1
MM1 pKa = 7.14SANRR5 pKa = 11.84ASTKK9 pKa = 10.06SVSRR13 pKa = 11.84RR14 pKa = 11.84PVAPRR19 pKa = 11.84QVVITTRR26 pKa = 11.84NAPRR30 pKa = 11.84AQKK33 pKa = 10.34SPKK36 pKa = 9.74KK37 pKa = 8.56PTPAKK42 pKa = 10.26EE43 pKa = 3.96AGFDD47 pKa = 3.85LNKK50 pKa = 9.98VVSDD54 pKa = 3.7ISSTFLAGLNKK65 pKa = 9.96PLVLLTLVLVVALVFTHH82 pKa = 6.6VNDD85 pKa = 4.82FSSGVLGKK93 pKa = 9.21WVAEE97 pKa = 4.01KK98 pKa = 10.69SANNSLALWVHH109 pKa = 4.98QNQMKK114 pKa = 10.3FLGLAIFAPAVLDD127 pKa = 3.86SPDD130 pKa = 4.18KK131 pKa = 10.87IRR133 pKa = 11.84LALALACVFWVMLVPQASVYY153 pKa = 10.23EE154 pKa = 4.09YY155 pKa = 10.46AIQAIALHH163 pKa = 6.07TYY165 pKa = 9.98FRR167 pKa = 11.84VRR169 pKa = 11.84LQNSRR174 pKa = 11.84VFILLLVAALYY185 pKa = 9.91FFGHH189 pKa = 6.19ITLSGTPGGTPVTNSSIGGG208 pKa = 3.59
MM1 pKa = 7.14SANRR5 pKa = 11.84ASTKK9 pKa = 10.06SVSRR13 pKa = 11.84RR14 pKa = 11.84PVAPRR19 pKa = 11.84QVVITTRR26 pKa = 11.84NAPRR30 pKa = 11.84AQKK33 pKa = 10.34SPKK36 pKa = 9.74KK37 pKa = 8.56PTPAKK42 pKa = 10.26EE43 pKa = 3.96AGFDD47 pKa = 3.85LNKK50 pKa = 9.98VVSDD54 pKa = 3.7ISSTFLAGLNKK65 pKa = 9.96PLVLLTLVLVVALVFTHH82 pKa = 6.6VNDD85 pKa = 4.82FSSGVLGKK93 pKa = 9.21WVAEE97 pKa = 4.01KK98 pKa = 10.69SANNSLALWVHH109 pKa = 4.98QNQMKK114 pKa = 10.3FLGLAIFAPAVLDD127 pKa = 3.86SPDD130 pKa = 4.18KK131 pKa = 10.87IRR133 pKa = 11.84LALALACVFWVMLVPQASVYY153 pKa = 10.23EE154 pKa = 4.09YY155 pKa = 10.46AIQAIALHH163 pKa = 6.07TYY165 pKa = 9.98FRR167 pKa = 11.84VRR169 pKa = 11.84LQNSRR174 pKa = 11.84VFILLLVAALYY185 pKa = 9.91FFGHH189 pKa = 6.19ITLSGTPGGTPVTNSSIGGG208 pKa = 3.59
Molecular weight: 22.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2971 |
208 |
2361 |
990.3 |
112.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.89 ± 1.347 | 2.02 ± 0.406 |
6.193 ± 1.121 | 5.419 ± 1.113 |
6.429 ± 0.569 | 4.645 ± 0.182 |
2.423 ± 0.24 | 5.453 ± 0.563 |
5.587 ± 0.26 | 8.852 ± 0.967 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.221 ± 0.25 | 4.948 ± 0.448 |
4.746 ± 0.599 | 2.726 ± 0.315 |
5.756 ± 0.768 | 8.078 ± 0.317 |
6.16 ± 0.219 | 8.112 ± 0.908 |
0.606 ± 0.304 | 3.736 ± 0.657 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
