
Penicillium janczewskii chrysovirus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Chrysoviridae; Betachrysovirus
Average proteome isoelectric point is 5.88
Get precalculated fractions of proteins
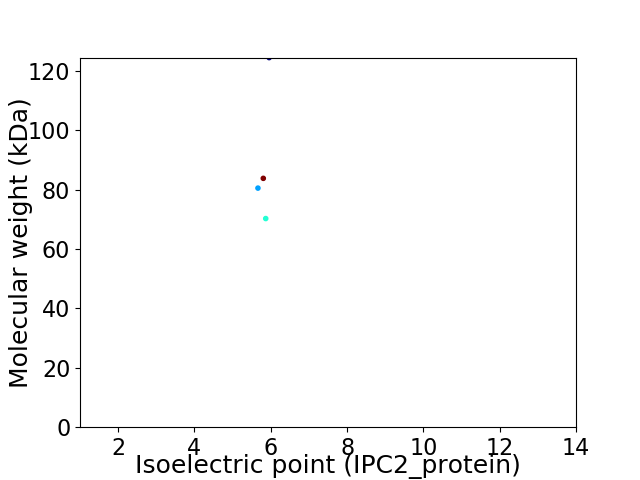
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
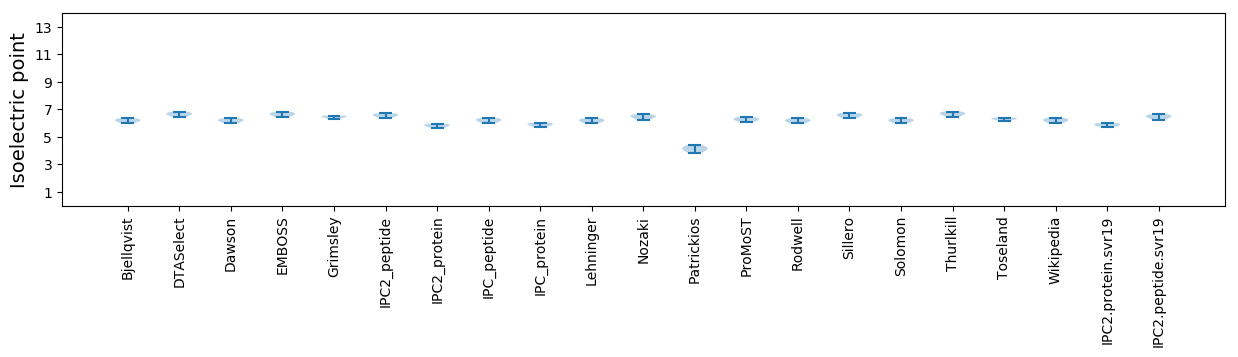
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0S2KPW5|A0A0S2KPW5_9VIRU RNA-directed RNA polymerase OS=Penicillium janczewskii chrysovirus 2 OX=1755793 PE=4 SV=1
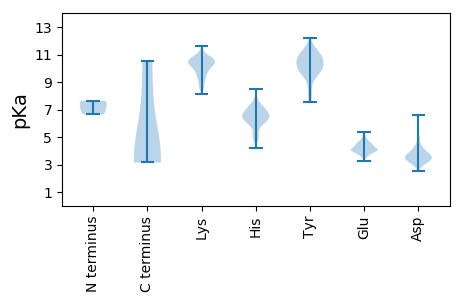
MM1 pKa = 7.63SSPLASPSAILGRR14 pKa = 11.84DD15 pKa = 3.53VPSLISSWADD25 pKa = 3.19EE26 pKa = 4.26VEE28 pKa = 3.86EE29 pKa = 4.95AYY31 pKa = 10.54ASGEE35 pKa = 4.05MATVEE40 pKa = 4.55DD41 pKa = 4.11GRR43 pKa = 11.84GAAGAASLVRR53 pKa = 11.84ADD55 pKa = 5.12AILSGTTAWGPEE67 pKa = 3.69SAARR71 pKa = 11.84SGLGATTLARR81 pKa = 11.84ATGLRR86 pKa = 11.84RR87 pKa = 11.84TTGAVFPPEE96 pKa = 4.56CEE98 pKa = 4.62DD99 pKa = 3.62GGHH102 pKa = 6.81LCGIPFGPRR111 pKa = 11.84VFAATWVDD119 pKa = 2.88ADD121 pKa = 3.32IAYY124 pKa = 10.02GSSEE128 pKa = 3.7PMRR131 pKa = 11.84RR132 pKa = 11.84CSDD135 pKa = 3.18NAIRR139 pKa = 11.84SLLRR143 pKa = 11.84TLDD146 pKa = 3.5GSEE149 pKa = 4.3GAFTLPSEE157 pKa = 4.68TRR159 pKa = 11.84CQVYY163 pKa = 10.14RR164 pKa = 11.84SMDD167 pKa = 2.98QHH169 pKa = 6.67MYY171 pKa = 9.99LQPDD175 pKa = 3.36EE176 pKa = 4.66GFFWVTFQVRR186 pKa = 11.84VEE188 pKa = 4.07AQLMVYY194 pKa = 7.91GHH196 pKa = 7.38PDD198 pKa = 2.74GRR200 pKa = 11.84IRR202 pKa = 11.84DD203 pKa = 3.72VAVARR208 pKa = 11.84DD209 pKa = 3.72LRR211 pKa = 11.84AMRR214 pKa = 11.84SADD217 pKa = 3.08VSAYY221 pKa = 10.45RR222 pKa = 11.84DD223 pKa = 3.74MLVVLAGIQQAIVFGEE239 pKa = 4.44LGCVLTVLHH248 pKa = 7.13DD249 pKa = 4.11CPLGEE254 pKa = 4.54IKK256 pKa = 10.56QSVHH260 pKa = 4.89RR261 pKa = 11.84HH262 pKa = 5.18PGGGVVTAGHH272 pKa = 6.51TGSPVQILDD281 pKa = 3.94TSGTWLPTPAQLHH294 pKa = 5.3HH295 pKa = 6.34QRR297 pKa = 11.84VSAATSFPEE306 pKa = 3.97GTVAHH311 pKa = 6.07GVVSASEE318 pKa = 4.04VMPTLVVGLGANGGKK333 pKa = 9.78SLTGSVLCVRR343 pKa = 11.84VLEE346 pKa = 4.64AGFAGARR353 pKa = 11.84NLVTTSTTANAPLPMFKK370 pKa = 9.71RR371 pKa = 11.84GMALAPCARR380 pKa = 11.84DD381 pKa = 3.09GLLSHH386 pKa = 6.91TCAAALYY393 pKa = 9.08GDD395 pKa = 3.78GLGGVFVYY403 pKa = 10.58PDD405 pKa = 3.79GVRR408 pKa = 11.84VAVDD412 pKa = 3.44TVLSKK417 pKa = 10.88LRR419 pKa = 11.84SDD421 pKa = 3.34GHH423 pKa = 6.5AWVYY427 pKa = 11.07ARR429 pKa = 11.84DD430 pKa = 3.79VVATKK435 pKa = 10.37QYY437 pKa = 10.9VLDD440 pKa = 5.63LISLQQGWDD449 pKa = 3.3RR450 pKa = 11.84QLDD453 pKa = 3.56NDD455 pKa = 4.37GAITVLDD462 pKa = 3.74SRR464 pKa = 11.84RR465 pKa = 11.84EE466 pKa = 4.18IIGSVYY472 pKa = 10.24VLPGAGSQPRR482 pKa = 11.84WATVLEE488 pKa = 4.73GVTMPTKK495 pKa = 10.47CGSEE499 pKa = 3.74LWLDD503 pKa = 3.65HH504 pKa = 6.27GVSGLGRR511 pKa = 11.84YY512 pKa = 8.89LLANHH517 pKa = 6.33ITNGRR522 pKa = 11.84FTSALSRR529 pKa = 11.84LAFNWKK535 pKa = 10.36GPFQASTAAVSAAKK549 pKa = 9.48TLGVSAGLQLPISYY563 pKa = 10.45VVDD566 pKa = 3.36TEE568 pKa = 4.47FISRR572 pKa = 11.84KK573 pKa = 10.23GPDD576 pKa = 3.66GNLDD580 pKa = 3.3RR581 pKa = 11.84YY582 pKa = 9.82VFAVGVARR590 pKa = 11.84FHH592 pKa = 7.34DD593 pKa = 4.35GQYY596 pKa = 9.4QGSMTVVDD604 pKa = 4.75TSAEE608 pKa = 4.0FHH610 pKa = 6.53EE611 pKa = 5.0FAAANRR617 pKa = 11.84SKK619 pKa = 10.75LGRR622 pKa = 11.84HH623 pKa = 4.19MASWEE628 pKa = 3.94VLQRR632 pKa = 11.84GAKK635 pKa = 10.01HH636 pKa = 6.3GDD638 pKa = 3.2PTVVLEE644 pKa = 4.19RR645 pKa = 11.84LRR647 pKa = 11.84EE648 pKa = 3.95VAADD652 pKa = 3.58TNVRR656 pKa = 11.84VYY658 pKa = 11.32AKK660 pKa = 10.44GYY662 pKa = 9.48DD663 pKa = 3.62AEE665 pKa = 4.57SEE667 pKa = 4.25ILVSEE672 pKa = 4.17PVGATRR678 pKa = 11.84LFRR681 pKa = 11.84RR682 pKa = 11.84QAGNRR687 pKa = 11.84APLRR691 pKa = 11.84EE692 pKa = 4.42LGLLTRR698 pKa = 11.84GYY700 pKa = 11.04DD701 pKa = 3.37EE702 pKa = 4.64LARR705 pKa = 11.84EE706 pKa = 4.36IGWPTDD712 pKa = 3.28HH713 pKa = 7.9DD714 pKa = 4.14PGRR717 pKa = 11.84EE718 pKa = 4.05CTLFGYY724 pKa = 7.7TAGLCDD730 pKa = 3.92SLPPLRR736 pKa = 11.84EE737 pKa = 3.71IGNDD741 pKa = 3.5SIHH744 pKa = 6.69AVLGSMSQAGILL756 pKa = 3.62
MM1 pKa = 7.63SSPLASPSAILGRR14 pKa = 11.84DD15 pKa = 3.53VPSLISSWADD25 pKa = 3.19EE26 pKa = 4.26VEE28 pKa = 3.86EE29 pKa = 4.95AYY31 pKa = 10.54ASGEE35 pKa = 4.05MATVEE40 pKa = 4.55DD41 pKa = 4.11GRR43 pKa = 11.84GAAGAASLVRR53 pKa = 11.84ADD55 pKa = 5.12AILSGTTAWGPEE67 pKa = 3.69SAARR71 pKa = 11.84SGLGATTLARR81 pKa = 11.84ATGLRR86 pKa = 11.84RR87 pKa = 11.84TTGAVFPPEE96 pKa = 4.56CEE98 pKa = 4.62DD99 pKa = 3.62GGHH102 pKa = 6.81LCGIPFGPRR111 pKa = 11.84VFAATWVDD119 pKa = 2.88ADD121 pKa = 3.32IAYY124 pKa = 10.02GSSEE128 pKa = 3.7PMRR131 pKa = 11.84RR132 pKa = 11.84CSDD135 pKa = 3.18NAIRR139 pKa = 11.84SLLRR143 pKa = 11.84TLDD146 pKa = 3.5GSEE149 pKa = 4.3GAFTLPSEE157 pKa = 4.68TRR159 pKa = 11.84CQVYY163 pKa = 10.14RR164 pKa = 11.84SMDD167 pKa = 2.98QHH169 pKa = 6.67MYY171 pKa = 9.99LQPDD175 pKa = 3.36EE176 pKa = 4.66GFFWVTFQVRR186 pKa = 11.84VEE188 pKa = 4.07AQLMVYY194 pKa = 7.91GHH196 pKa = 7.38PDD198 pKa = 2.74GRR200 pKa = 11.84IRR202 pKa = 11.84DD203 pKa = 3.72VAVARR208 pKa = 11.84DD209 pKa = 3.72LRR211 pKa = 11.84AMRR214 pKa = 11.84SADD217 pKa = 3.08VSAYY221 pKa = 10.45RR222 pKa = 11.84DD223 pKa = 3.74MLVVLAGIQQAIVFGEE239 pKa = 4.44LGCVLTVLHH248 pKa = 7.13DD249 pKa = 4.11CPLGEE254 pKa = 4.54IKK256 pKa = 10.56QSVHH260 pKa = 4.89RR261 pKa = 11.84HH262 pKa = 5.18PGGGVVTAGHH272 pKa = 6.51TGSPVQILDD281 pKa = 3.94TSGTWLPTPAQLHH294 pKa = 5.3HH295 pKa = 6.34QRR297 pKa = 11.84VSAATSFPEE306 pKa = 3.97GTVAHH311 pKa = 6.07GVVSASEE318 pKa = 4.04VMPTLVVGLGANGGKK333 pKa = 9.78SLTGSVLCVRR343 pKa = 11.84VLEE346 pKa = 4.64AGFAGARR353 pKa = 11.84NLVTTSTTANAPLPMFKK370 pKa = 9.71RR371 pKa = 11.84GMALAPCARR380 pKa = 11.84DD381 pKa = 3.09GLLSHH386 pKa = 6.91TCAAALYY393 pKa = 9.08GDD395 pKa = 3.78GLGGVFVYY403 pKa = 10.58PDD405 pKa = 3.79GVRR408 pKa = 11.84VAVDD412 pKa = 3.44TVLSKK417 pKa = 10.88LRR419 pKa = 11.84SDD421 pKa = 3.34GHH423 pKa = 6.5AWVYY427 pKa = 11.07ARR429 pKa = 11.84DD430 pKa = 3.79VVATKK435 pKa = 10.37QYY437 pKa = 10.9VLDD440 pKa = 5.63LISLQQGWDD449 pKa = 3.3RR450 pKa = 11.84QLDD453 pKa = 3.56NDD455 pKa = 4.37GAITVLDD462 pKa = 3.74SRR464 pKa = 11.84RR465 pKa = 11.84EE466 pKa = 4.18IIGSVYY472 pKa = 10.24VLPGAGSQPRR482 pKa = 11.84WATVLEE488 pKa = 4.73GVTMPTKK495 pKa = 10.47CGSEE499 pKa = 3.74LWLDD503 pKa = 3.65HH504 pKa = 6.27GVSGLGRR511 pKa = 11.84YY512 pKa = 8.89LLANHH517 pKa = 6.33ITNGRR522 pKa = 11.84FTSALSRR529 pKa = 11.84LAFNWKK535 pKa = 10.36GPFQASTAAVSAAKK549 pKa = 9.48TLGVSAGLQLPISYY563 pKa = 10.45VVDD566 pKa = 3.36TEE568 pKa = 4.47FISRR572 pKa = 11.84KK573 pKa = 10.23GPDD576 pKa = 3.66GNLDD580 pKa = 3.3RR581 pKa = 11.84YY582 pKa = 9.82VFAVGVARR590 pKa = 11.84FHH592 pKa = 7.34DD593 pKa = 4.35GQYY596 pKa = 9.4QGSMTVVDD604 pKa = 4.75TSAEE608 pKa = 4.0FHH610 pKa = 6.53EE611 pKa = 5.0FAAANRR617 pKa = 11.84SKK619 pKa = 10.75LGRR622 pKa = 11.84HH623 pKa = 4.19MASWEE628 pKa = 3.94VLQRR632 pKa = 11.84GAKK635 pKa = 10.01HH636 pKa = 6.3GDD638 pKa = 3.2PTVVLEE644 pKa = 4.19RR645 pKa = 11.84LRR647 pKa = 11.84EE648 pKa = 3.95VAADD652 pKa = 3.58TNVRR656 pKa = 11.84VYY658 pKa = 11.32AKK660 pKa = 10.44GYY662 pKa = 9.48DD663 pKa = 3.62AEE665 pKa = 4.57SEE667 pKa = 4.25ILVSEE672 pKa = 4.17PVGATRR678 pKa = 11.84LFRR681 pKa = 11.84RR682 pKa = 11.84QAGNRR687 pKa = 11.84APLRR691 pKa = 11.84EE692 pKa = 4.42LGLLTRR698 pKa = 11.84GYY700 pKa = 11.04DD701 pKa = 3.37EE702 pKa = 4.64LARR705 pKa = 11.84EE706 pKa = 4.36IGWPTDD712 pKa = 3.28HH713 pKa = 7.9DD714 pKa = 4.14PGRR717 pKa = 11.84EE718 pKa = 4.05CTLFGYY724 pKa = 7.7TAGLCDD730 pKa = 3.92SLPPLRR736 pKa = 11.84EE737 pKa = 3.71IGNDD741 pKa = 3.5SIHH744 pKa = 6.69AVLGSMSQAGILL756 pKa = 3.62
Molecular weight: 80.49 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0S2KQ51|A0A0S2KQ51_9VIRU ORF2 OS=Penicillium janczewskii chrysovirus 2 OX=1755793 PE=4 SV=1
MM1 pKa = 6.96NTPTLGASYY10 pKa = 10.09TRR12 pKa = 11.84TPTGGNSTRR21 pKa = 11.84LFASDD26 pKa = 3.62NDD28 pKa = 3.86AKK30 pKa = 10.88LKK32 pKa = 10.0EE33 pKa = 4.33WEE35 pKa = 4.18GLHH38 pKa = 7.16AIVLPTCCGKK48 pKa = 10.69SSAALRR54 pKa = 11.84FGGYY58 pKa = 10.45DD59 pKa = 3.01IDD61 pKa = 6.58DD62 pKa = 3.86IVADD66 pKa = 4.17ASIDD70 pKa = 3.84EE71 pKa = 5.36LDD73 pKa = 3.92TEE75 pKa = 4.68LDD77 pKa = 3.26AMLGARR83 pKa = 11.84EE84 pKa = 4.51DD85 pKa = 4.18GVHH88 pKa = 6.48GNDD91 pKa = 2.92SSAMHH96 pKa = 6.76RR97 pKa = 11.84SNRR100 pKa = 11.84LMLMRR105 pKa = 11.84ARR107 pKa = 11.84RR108 pKa = 11.84FFATVSPDD116 pKa = 3.54DD117 pKa = 3.95NPLVVYY123 pKa = 9.53CHH125 pKa = 5.77TAEE128 pKa = 4.13FAQALGVPVGLVVHH142 pKa = 6.88LDD144 pKa = 3.54EE145 pKa = 4.98EE146 pKa = 4.69AVSNSIRR153 pKa = 11.84MTRR156 pKa = 11.84EE157 pKa = 3.27TTPEE161 pKa = 3.71VRR163 pKa = 11.84NVTLQVYY170 pKa = 9.54RR171 pKa = 11.84EE172 pKa = 3.8QTAANRR178 pKa = 11.84EE179 pKa = 3.88FSRR182 pKa = 11.84RR183 pKa = 11.84HH184 pKa = 4.97GLSSRR189 pKa = 11.84DD190 pKa = 3.59CYY192 pKa = 11.15SYY194 pKa = 11.67SDD196 pKa = 3.68VQQCVWSALVRR207 pKa = 11.84CAVLAPCRR215 pKa = 11.84QGDD218 pKa = 4.18EE219 pKa = 4.1YY220 pKa = 11.33HH221 pKa = 7.08AMLDD225 pKa = 3.95SRR227 pKa = 11.84PQLSDD232 pKa = 3.11VLNRR236 pKa = 11.84SHH238 pKa = 6.91AVVRR242 pKa = 11.84TTSTPNWVRR251 pKa = 11.84ACAARR256 pKa = 11.84QLKK259 pKa = 10.62LSLGDD264 pKa = 3.83LAPTEE269 pKa = 3.79AHH271 pKa = 6.74AAHH274 pKa = 6.64SHH276 pKa = 6.0PAWASIVHH284 pKa = 6.15IVASQTGHH292 pKa = 5.95VPEE295 pKa = 4.13QAYY298 pKa = 10.54ASIPDD303 pKa = 3.34WSEE306 pKa = 3.99EE307 pKa = 3.54EE308 pKa = 3.72WRR310 pKa = 11.84EE311 pKa = 3.91YY312 pKa = 10.99FPLGPGNASFALVNVSDD329 pKa = 3.76WLARR333 pKa = 11.84TPSDD337 pKa = 3.22ALRR340 pKa = 11.84STAYY344 pKa = 9.88SWFRR348 pKa = 11.84QLLLVRR354 pKa = 11.84GTKK357 pKa = 8.56YY358 pKa = 10.51EE359 pKa = 4.09RR360 pKa = 11.84LLCMLVMGDD369 pKa = 3.33VSHH372 pKa = 6.58YY373 pKa = 9.95VAPQYY378 pKa = 10.77TEE380 pKa = 4.14LYY382 pKa = 10.41SRR384 pKa = 11.84IPLGLLSDD392 pKa = 3.82VVYY395 pKa = 9.18ATISKK400 pKa = 9.85QIHH403 pKa = 5.09AAVRR407 pKa = 11.84VGCNYY412 pKa = 10.26LGQRR416 pKa = 11.84IPSRR420 pKa = 11.84DD421 pKa = 3.35LTYY424 pKa = 11.0FMYY427 pKa = 10.45FDD429 pKa = 4.35CLAGRR434 pKa = 11.84VIGTEE439 pKa = 4.49DD440 pKa = 3.41IDD442 pKa = 5.26AEE444 pKa = 4.17IADD447 pKa = 4.41RR448 pKa = 11.84TRR450 pKa = 11.84LQSPKK455 pKa = 9.94KK456 pKa = 9.48FCLGGEE462 pKa = 3.98WSEE465 pKa = 4.81SEE467 pKa = 3.65FDD469 pKa = 3.63NRR471 pKa = 11.84FTGAVQKK478 pKa = 10.81AYY480 pKa = 10.89DD481 pKa = 4.33HH482 pKa = 6.27LTQGLGAKK490 pKa = 9.16LRR492 pKa = 11.84AMTNVIDD499 pKa = 4.14TFDD502 pKa = 3.27HH503 pKa = 6.2FMEE506 pKa = 4.79EE507 pKa = 3.3RR508 pKa = 11.84RR509 pKa = 11.84TWVRR513 pKa = 11.84PGAASGAPKK522 pKa = 10.18ADD524 pKa = 3.49LMLEE528 pKa = 4.47VPASQAEE535 pKa = 4.06ALDD538 pKa = 5.01AISTDD543 pKa = 3.6LGHH546 pKa = 5.74MTVLILRR553 pKa = 11.84RR554 pKa = 11.84VRR556 pKa = 11.84LNKK559 pKa = 10.09SAVFEE564 pKa = 4.3FPQFVDD570 pKa = 3.8TVKK573 pKa = 10.57EE574 pKa = 3.82ALRR577 pKa = 11.84DD578 pKa = 3.67YY579 pKa = 10.92VPNSFTKK586 pKa = 10.69YY587 pKa = 9.59FFKK590 pKa = 11.0FEE592 pKa = 4.44PGKK595 pKa = 9.95FASRR599 pKa = 11.84ALFPSHH605 pKa = 6.95LLHH608 pKa = 7.26YY609 pKa = 10.71IMVSHH614 pKa = 6.42VLFVAEE620 pKa = 4.37KK621 pKa = 10.26GGVIPEE627 pKa = 4.22TRR629 pKa = 11.84LTAGPEE635 pKa = 4.05AQQEE639 pKa = 4.29DD640 pKa = 3.52HH641 pKa = 6.22WLWRR645 pKa = 11.84EE646 pKa = 3.71MHH648 pKa = 6.49DD649 pKa = 3.82TAVHH653 pKa = 6.79LMLDD657 pKa = 3.45YY658 pKa = 11.66ANFNEE663 pKa = 4.21QHH665 pKa = 6.41SIKK668 pKa = 10.48HH669 pKa = 4.61MQATILGLKK678 pKa = 9.58GVYY681 pKa = 9.66SKK683 pKa = 10.88HH684 pKa = 6.81LSLTPDD690 pKa = 4.02LARR693 pKa = 11.84AIDD696 pKa = 3.53WTVEE700 pKa = 3.81SFEE703 pKa = 4.92RR704 pKa = 11.84ICAEE708 pKa = 3.58QDD710 pKa = 2.97GKK712 pKa = 10.69LVRR715 pKa = 11.84FTHH718 pKa = 6.49GLLSGWRR725 pKa = 11.84CTTWVNSIANVAYY738 pKa = 10.59LDD740 pKa = 4.05VIRR743 pKa = 11.84QQVQEE748 pKa = 3.94LTGRR752 pKa = 11.84SVLVRR757 pKa = 11.84TQSGGDD763 pKa = 3.27DD764 pKa = 3.49VAAEE768 pKa = 4.17EE769 pKa = 4.57CSLYY773 pKa = 10.93DD774 pKa = 3.14AALTLRR780 pKa = 11.84VGEE783 pKa = 4.12VMGFEE788 pKa = 4.67FKK790 pKa = 10.58AIKK793 pKa = 10.04QLISCEE799 pKa = 3.65YY800 pKa = 10.47RR801 pKa = 11.84EE802 pKa = 4.13FFRR805 pKa = 11.84LFITRR810 pKa = 11.84EE811 pKa = 3.81GVYY814 pKa = 10.89GSLCRR819 pKa = 11.84MLGSALSGQWSNSVLPKK836 pKa = 10.32LVEE839 pKa = 4.16PATKK843 pKa = 10.21LSSVVEE849 pKa = 4.19IARR852 pKa = 11.84KK853 pKa = 8.86AGRR856 pKa = 11.84RR857 pKa = 11.84ARR859 pKa = 11.84NLNLMEE865 pKa = 4.58KK866 pKa = 8.67MALVAFDD873 pKa = 3.41KK874 pKa = 10.57WATTGEE880 pKa = 4.06EE881 pKa = 4.37CLVDD885 pKa = 4.88FYY887 pKa = 12.21VHH889 pKa = 5.73GTKK892 pKa = 8.79EE893 pKa = 4.21TGGLGIPNVTGDD905 pKa = 4.44LYY907 pKa = 11.76VLDD910 pKa = 4.99GSPDD914 pKa = 3.32EE915 pKa = 5.31RR916 pKa = 11.84PTHH919 pKa = 6.28LKK921 pKa = 10.33PVGHH925 pKa = 7.25PSDD928 pKa = 3.89ASSVTAARR936 pKa = 11.84IVDD939 pKa = 3.73KK940 pKa = 10.79AVSLVGPEE948 pKa = 4.05AVLPTATVASTMAEE962 pKa = 4.11GAFVSSIAQNLGPRR976 pKa = 11.84ALRR979 pKa = 11.84VGGRR983 pKa = 11.84RR984 pKa = 11.84QSHH987 pKa = 5.06KK988 pKa = 9.55VVRR991 pKa = 11.84VLRR994 pKa = 11.84IRR996 pKa = 11.84EE997 pKa = 3.77EE998 pKa = 4.02DD999 pKa = 3.63VAPGRR1004 pKa = 11.84TSADD1008 pKa = 3.53YY1009 pKa = 10.6QHH1011 pKa = 6.84SKK1013 pKa = 10.4LALRR1017 pKa = 11.84VQLDD1021 pKa = 3.17AMKK1024 pKa = 10.55RR1025 pKa = 11.84AGRR1028 pKa = 11.84RR1029 pKa = 11.84YY1030 pKa = 10.38AEE1032 pKa = 4.49LAPAVKK1038 pKa = 9.24PAKK1041 pKa = 10.28KK1042 pKa = 9.98LDD1044 pKa = 3.56LARR1047 pKa = 11.84SVGASEE1053 pKa = 5.12GVGGDD1058 pKa = 3.87LLYY1061 pKa = 10.78YY1062 pKa = 8.6WQEE1065 pKa = 3.56EE1066 pKa = 4.3HH1067 pKa = 6.88VLYY1070 pKa = 11.07GCATYY1075 pKa = 10.25MLTEE1079 pKa = 5.05DD1080 pKa = 4.0YY1081 pKa = 11.29YY1082 pKa = 11.31EE1083 pKa = 4.37DD1084 pKa = 3.6VVLIGLLQRR1093 pKa = 11.84GRR1095 pKa = 11.84DD1096 pKa = 3.27RR1097 pKa = 11.84CAVANRR1103 pKa = 11.84CAEE1106 pKa = 4.02LAMGLRR1112 pKa = 11.84NDD1114 pKa = 3.47GYY1116 pKa = 9.79MWYY1119 pKa = 10.56
MM1 pKa = 6.96NTPTLGASYY10 pKa = 10.09TRR12 pKa = 11.84TPTGGNSTRR21 pKa = 11.84LFASDD26 pKa = 3.62NDD28 pKa = 3.86AKK30 pKa = 10.88LKK32 pKa = 10.0EE33 pKa = 4.33WEE35 pKa = 4.18GLHH38 pKa = 7.16AIVLPTCCGKK48 pKa = 10.69SSAALRR54 pKa = 11.84FGGYY58 pKa = 10.45DD59 pKa = 3.01IDD61 pKa = 6.58DD62 pKa = 3.86IVADD66 pKa = 4.17ASIDD70 pKa = 3.84EE71 pKa = 5.36LDD73 pKa = 3.92TEE75 pKa = 4.68LDD77 pKa = 3.26AMLGARR83 pKa = 11.84EE84 pKa = 4.51DD85 pKa = 4.18GVHH88 pKa = 6.48GNDD91 pKa = 2.92SSAMHH96 pKa = 6.76RR97 pKa = 11.84SNRR100 pKa = 11.84LMLMRR105 pKa = 11.84ARR107 pKa = 11.84RR108 pKa = 11.84FFATVSPDD116 pKa = 3.54DD117 pKa = 3.95NPLVVYY123 pKa = 9.53CHH125 pKa = 5.77TAEE128 pKa = 4.13FAQALGVPVGLVVHH142 pKa = 6.88LDD144 pKa = 3.54EE145 pKa = 4.98EE146 pKa = 4.69AVSNSIRR153 pKa = 11.84MTRR156 pKa = 11.84EE157 pKa = 3.27TTPEE161 pKa = 3.71VRR163 pKa = 11.84NVTLQVYY170 pKa = 9.54RR171 pKa = 11.84EE172 pKa = 3.8QTAANRR178 pKa = 11.84EE179 pKa = 3.88FSRR182 pKa = 11.84RR183 pKa = 11.84HH184 pKa = 4.97GLSSRR189 pKa = 11.84DD190 pKa = 3.59CYY192 pKa = 11.15SYY194 pKa = 11.67SDD196 pKa = 3.68VQQCVWSALVRR207 pKa = 11.84CAVLAPCRR215 pKa = 11.84QGDD218 pKa = 4.18EE219 pKa = 4.1YY220 pKa = 11.33HH221 pKa = 7.08AMLDD225 pKa = 3.95SRR227 pKa = 11.84PQLSDD232 pKa = 3.11VLNRR236 pKa = 11.84SHH238 pKa = 6.91AVVRR242 pKa = 11.84TTSTPNWVRR251 pKa = 11.84ACAARR256 pKa = 11.84QLKK259 pKa = 10.62LSLGDD264 pKa = 3.83LAPTEE269 pKa = 3.79AHH271 pKa = 6.74AAHH274 pKa = 6.64SHH276 pKa = 6.0PAWASIVHH284 pKa = 6.15IVASQTGHH292 pKa = 5.95VPEE295 pKa = 4.13QAYY298 pKa = 10.54ASIPDD303 pKa = 3.34WSEE306 pKa = 3.99EE307 pKa = 3.54EE308 pKa = 3.72WRR310 pKa = 11.84EE311 pKa = 3.91YY312 pKa = 10.99FPLGPGNASFALVNVSDD329 pKa = 3.76WLARR333 pKa = 11.84TPSDD337 pKa = 3.22ALRR340 pKa = 11.84STAYY344 pKa = 9.88SWFRR348 pKa = 11.84QLLLVRR354 pKa = 11.84GTKK357 pKa = 8.56YY358 pKa = 10.51EE359 pKa = 4.09RR360 pKa = 11.84LLCMLVMGDD369 pKa = 3.33VSHH372 pKa = 6.58YY373 pKa = 9.95VAPQYY378 pKa = 10.77TEE380 pKa = 4.14LYY382 pKa = 10.41SRR384 pKa = 11.84IPLGLLSDD392 pKa = 3.82VVYY395 pKa = 9.18ATISKK400 pKa = 9.85QIHH403 pKa = 5.09AAVRR407 pKa = 11.84VGCNYY412 pKa = 10.26LGQRR416 pKa = 11.84IPSRR420 pKa = 11.84DD421 pKa = 3.35LTYY424 pKa = 11.0FMYY427 pKa = 10.45FDD429 pKa = 4.35CLAGRR434 pKa = 11.84VIGTEE439 pKa = 4.49DD440 pKa = 3.41IDD442 pKa = 5.26AEE444 pKa = 4.17IADD447 pKa = 4.41RR448 pKa = 11.84TRR450 pKa = 11.84LQSPKK455 pKa = 9.94KK456 pKa = 9.48FCLGGEE462 pKa = 3.98WSEE465 pKa = 4.81SEE467 pKa = 3.65FDD469 pKa = 3.63NRR471 pKa = 11.84FTGAVQKK478 pKa = 10.81AYY480 pKa = 10.89DD481 pKa = 4.33HH482 pKa = 6.27LTQGLGAKK490 pKa = 9.16LRR492 pKa = 11.84AMTNVIDD499 pKa = 4.14TFDD502 pKa = 3.27HH503 pKa = 6.2FMEE506 pKa = 4.79EE507 pKa = 3.3RR508 pKa = 11.84RR509 pKa = 11.84TWVRR513 pKa = 11.84PGAASGAPKK522 pKa = 10.18ADD524 pKa = 3.49LMLEE528 pKa = 4.47VPASQAEE535 pKa = 4.06ALDD538 pKa = 5.01AISTDD543 pKa = 3.6LGHH546 pKa = 5.74MTVLILRR553 pKa = 11.84RR554 pKa = 11.84VRR556 pKa = 11.84LNKK559 pKa = 10.09SAVFEE564 pKa = 4.3FPQFVDD570 pKa = 3.8TVKK573 pKa = 10.57EE574 pKa = 3.82ALRR577 pKa = 11.84DD578 pKa = 3.67YY579 pKa = 10.92VPNSFTKK586 pKa = 10.69YY587 pKa = 9.59FFKK590 pKa = 11.0FEE592 pKa = 4.44PGKK595 pKa = 9.95FASRR599 pKa = 11.84ALFPSHH605 pKa = 6.95LLHH608 pKa = 7.26YY609 pKa = 10.71IMVSHH614 pKa = 6.42VLFVAEE620 pKa = 4.37KK621 pKa = 10.26GGVIPEE627 pKa = 4.22TRR629 pKa = 11.84LTAGPEE635 pKa = 4.05AQQEE639 pKa = 4.29DD640 pKa = 3.52HH641 pKa = 6.22WLWRR645 pKa = 11.84EE646 pKa = 3.71MHH648 pKa = 6.49DD649 pKa = 3.82TAVHH653 pKa = 6.79LMLDD657 pKa = 3.45YY658 pKa = 11.66ANFNEE663 pKa = 4.21QHH665 pKa = 6.41SIKK668 pKa = 10.48HH669 pKa = 4.61MQATILGLKK678 pKa = 9.58GVYY681 pKa = 9.66SKK683 pKa = 10.88HH684 pKa = 6.81LSLTPDD690 pKa = 4.02LARR693 pKa = 11.84AIDD696 pKa = 3.53WTVEE700 pKa = 3.81SFEE703 pKa = 4.92RR704 pKa = 11.84ICAEE708 pKa = 3.58QDD710 pKa = 2.97GKK712 pKa = 10.69LVRR715 pKa = 11.84FTHH718 pKa = 6.49GLLSGWRR725 pKa = 11.84CTTWVNSIANVAYY738 pKa = 10.59LDD740 pKa = 4.05VIRR743 pKa = 11.84QQVQEE748 pKa = 3.94LTGRR752 pKa = 11.84SVLVRR757 pKa = 11.84TQSGGDD763 pKa = 3.27DD764 pKa = 3.49VAAEE768 pKa = 4.17EE769 pKa = 4.57CSLYY773 pKa = 10.93DD774 pKa = 3.14AALTLRR780 pKa = 11.84VGEE783 pKa = 4.12VMGFEE788 pKa = 4.67FKK790 pKa = 10.58AIKK793 pKa = 10.04QLISCEE799 pKa = 3.65YY800 pKa = 10.47RR801 pKa = 11.84EE802 pKa = 4.13FFRR805 pKa = 11.84LFITRR810 pKa = 11.84EE811 pKa = 3.81GVYY814 pKa = 10.89GSLCRR819 pKa = 11.84MLGSALSGQWSNSVLPKK836 pKa = 10.32LVEE839 pKa = 4.16PATKK843 pKa = 10.21LSSVVEE849 pKa = 4.19IARR852 pKa = 11.84KK853 pKa = 8.86AGRR856 pKa = 11.84RR857 pKa = 11.84ARR859 pKa = 11.84NLNLMEE865 pKa = 4.58KK866 pKa = 8.67MALVAFDD873 pKa = 3.41KK874 pKa = 10.57WATTGEE880 pKa = 4.06EE881 pKa = 4.37CLVDD885 pKa = 4.88FYY887 pKa = 12.21VHH889 pKa = 5.73GTKK892 pKa = 8.79EE893 pKa = 4.21TGGLGIPNVTGDD905 pKa = 4.44LYY907 pKa = 11.76VLDD910 pKa = 4.99GSPDD914 pKa = 3.32EE915 pKa = 5.31RR916 pKa = 11.84PTHH919 pKa = 6.28LKK921 pKa = 10.33PVGHH925 pKa = 7.25PSDD928 pKa = 3.89ASSVTAARR936 pKa = 11.84IVDD939 pKa = 3.73KK940 pKa = 10.79AVSLVGPEE948 pKa = 4.05AVLPTATVASTMAEE962 pKa = 4.11GAFVSSIAQNLGPRR976 pKa = 11.84ALRR979 pKa = 11.84VGGRR983 pKa = 11.84RR984 pKa = 11.84QSHH987 pKa = 5.06KK988 pKa = 9.55VVRR991 pKa = 11.84VLRR994 pKa = 11.84IRR996 pKa = 11.84EE997 pKa = 3.77EE998 pKa = 4.02DD999 pKa = 3.63VAPGRR1004 pKa = 11.84TSADD1008 pKa = 3.53YY1009 pKa = 10.6QHH1011 pKa = 6.84SKK1013 pKa = 10.4LALRR1017 pKa = 11.84VQLDD1021 pKa = 3.17AMKK1024 pKa = 10.55RR1025 pKa = 11.84AGRR1028 pKa = 11.84RR1029 pKa = 11.84YY1030 pKa = 10.38AEE1032 pKa = 4.49LAPAVKK1038 pKa = 9.24PAKK1041 pKa = 10.28KK1042 pKa = 9.98LDD1044 pKa = 3.56LARR1047 pKa = 11.84SVGASEE1053 pKa = 5.12GVGGDD1058 pKa = 3.87LLYY1061 pKa = 10.78YY1062 pKa = 8.6WQEE1065 pKa = 3.56EE1066 pKa = 4.3HH1067 pKa = 6.88VLYY1070 pKa = 11.07GCATYY1075 pKa = 10.25MLTEE1079 pKa = 5.05DD1080 pKa = 4.0YY1081 pKa = 11.29YY1082 pKa = 11.31EE1083 pKa = 4.37DD1084 pKa = 3.6VVLIGLLQRR1093 pKa = 11.84GRR1095 pKa = 11.84DD1096 pKa = 3.27RR1097 pKa = 11.84CAVANRR1103 pKa = 11.84CAEE1106 pKa = 4.02LAMGLRR1112 pKa = 11.84NDD1114 pKa = 3.47GYY1116 pKa = 9.79MWYY1119 pKa = 10.56
Molecular weight: 124.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3313 |
655 |
1119 |
828.3 |
89.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.651 ± 0.537 | 1.57 ± 0.132 |
5.403 ± 0.233 | 5.343 ± 0.314 |
2.656 ± 0.286 | 8.391 ± 0.926 |
2.777 ± 0.13 | 3.049 ± 0.037 |
2.324 ± 0.384 | 9.81 ± 0.329 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.264 ± 0.122 | 2.203 ± 0.187 |
5.312 ± 0.639 | 3.441 ± 0.305 |
7.274 ± 0.167 | 7.486 ± 0.433 |
6.007 ± 0.186 | 8.421 ± 0.228 |
1.6 ± 0.056 | 3.018 ± 0.431 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
