
Bosea sp. Leaf344
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Boseaceae; Bosea; unclassified Bosea
Average proteome isoelectric point is 6.88
Get precalculated fractions of proteins
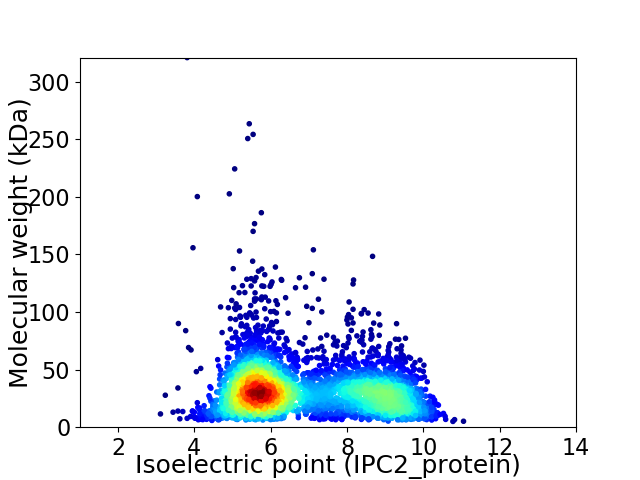
Virtual 2D-PAGE plot for 3876 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
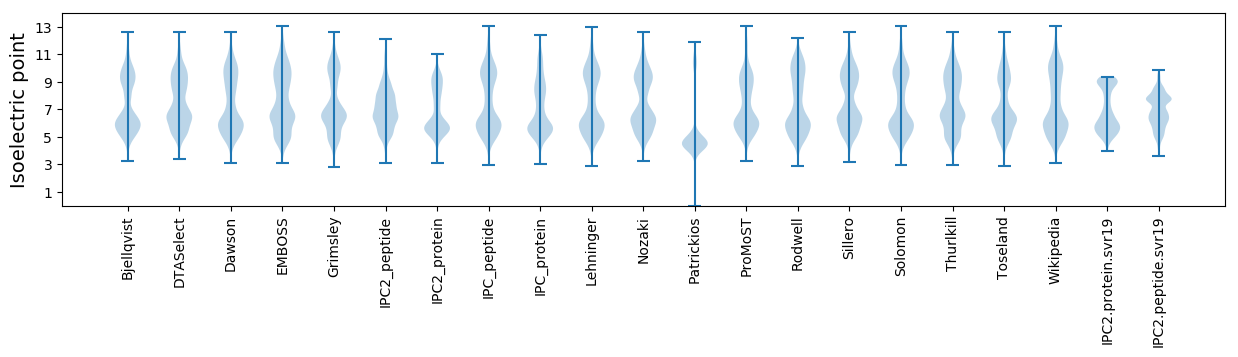
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q6KMY9|A0A0Q6KMY9_9BRAD 1-deoxy-D-xylulose-5-phosphate synthase OS=Bosea sp. Leaf344 OX=1736346 GN=dxs PE=3 SV=1
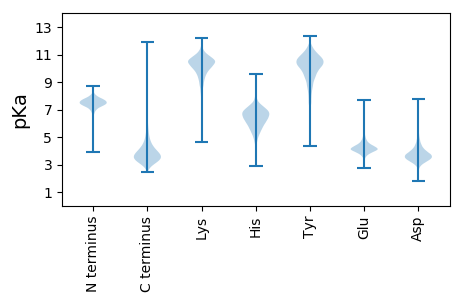
MM1 pKa = 7.18NVGGFSEE8 pKa = 5.17SIWHH12 pKa = 5.33FAGYY16 pKa = 10.15LRR18 pKa = 11.84LIEE21 pKa = 4.14PAARR25 pKa = 11.84PSDD28 pKa = 4.41LFDD31 pKa = 5.24GDD33 pKa = 4.49PLPPPGDD40 pKa = 3.77EE41 pKa = 3.78IYY43 pKa = 11.25GRR45 pKa = 11.84LSEE48 pKa = 4.6TLPPAQFEE56 pKa = 4.41EE57 pKa = 4.75TASQRR62 pKa = 11.84VLFVEE67 pKa = 5.27APAASPALPGLKK79 pKa = 9.8LASPDD84 pKa = 3.79FKK86 pKa = 10.85PISLNPSSFPALPRR100 pKa = 11.84RR101 pKa = 11.84DD102 pKa = 4.13APLQEE107 pKa = 4.38PDD109 pKa = 2.98IALEE113 pKa = 3.98GGIQFRR119 pKa = 11.84FLSQSRR125 pKa = 11.84SISVDD130 pKa = 3.34YY131 pKa = 11.04QPGGTEE137 pKa = 3.6TLLQLRR143 pKa = 11.84QINTMDD149 pKa = 3.95DD150 pKa = 3.5RR151 pKa = 11.84DD152 pKa = 4.34VILSDD157 pKa = 4.0ALPGVLPPEE166 pKa = 4.61IEE168 pKa = 3.89TGAAVAEE175 pKa = 4.33MLQQAQEE182 pKa = 4.13ATPDD186 pKa = 3.67SLPFEE191 pKa = 5.57AIGSTQALIAAITARR206 pKa = 11.84DD207 pKa = 3.7AAWAQSGQSPHH218 pKa = 7.19ADD220 pKa = 3.15PDD222 pKa = 3.7YY223 pKa = 10.21TPPSGLIVDD232 pKa = 4.6GVVVAAAPTGPTIEE246 pKa = 4.99QIAPWRR252 pKa = 11.84EE253 pKa = 3.61AASAPTQFVSQTVSEE268 pKa = 4.26AASTLGIGAVAEE280 pKa = 4.35TGLNQQINQAVILDD294 pKa = 3.94LNEE297 pKa = 4.45AVGSMIVGGDD307 pKa = 3.15AFFSRR312 pKa = 11.84GIVQVNILTDD322 pKa = 3.55SDD324 pKa = 4.05HH325 pKa = 8.05VDD327 pKa = 3.07IAIEE331 pKa = 4.18GAGSVGVFTTDD342 pKa = 3.13NQLHH346 pKa = 6.54NIAEE350 pKa = 4.49FVTHH354 pKa = 7.46DD355 pKa = 3.08MTARR359 pKa = 11.84YY360 pKa = 9.24SGAAATPYY368 pKa = 9.49WHH370 pKa = 7.27VDD372 pKa = 3.11TLAGNFYY379 pKa = 10.6DD380 pKa = 4.91VKK382 pKa = 10.54TLVQFNGLEE391 pKa = 4.35DD392 pKa = 3.86GDD394 pKa = 3.87RR395 pKa = 11.84TVQASEE401 pKa = 3.84GAYY404 pKa = 10.36FEE406 pKa = 5.6AKK408 pKa = 9.51TGLNGQVNYY417 pKa = 11.45AMVTSLDD424 pKa = 3.4QYY426 pKa = 11.33DD427 pKa = 4.05IIIIAGDD434 pKa = 3.3YY435 pKa = 10.6HH436 pKa = 6.17RR437 pKa = 11.84ADD439 pKa = 3.93WIFQYY444 pKa = 11.34NIMLDD449 pKa = 3.39SDD451 pKa = 4.07VAKK454 pKa = 10.58LYY456 pKa = 10.78AAGGTAEE463 pKa = 5.39DD464 pKa = 4.59DD465 pKa = 3.99VAVTTGFNSLTNSASITTYY484 pKa = 11.21DD485 pKa = 3.26SAAFAALNAAQHH497 pKa = 6.65DD498 pKa = 4.75LLSALAQGATVLTPHH513 pKa = 7.29ADD515 pKa = 2.69WGLNGNISGEE525 pKa = 4.09LKK527 pKa = 10.15ILYY530 pKa = 10.18VSGDD534 pKa = 3.8YY535 pKa = 11.27YY536 pKa = 11.28DD537 pKa = 4.3VNVITQINLMVDD549 pKa = 3.52ADD551 pKa = 3.82QAIQASAIGGEE562 pKa = 4.26LGVAAGGNSAANYY575 pKa = 10.66AEE577 pKa = 4.81IIDD580 pKa = 4.79PGTLSTSRR588 pKa = 11.84YY589 pKa = 8.96IGGEE593 pKa = 4.0AYY595 pKa = 10.44DD596 pKa = 4.44DD597 pKa = 5.19AILVQTNLVTDD608 pKa = 4.54DD609 pKa = 4.06DD610 pKa = 4.68TVTIHH615 pKa = 7.14DD616 pKa = 4.08TTTLVPEE623 pKa = 4.75FVAFAEE629 pKa = 4.27QTEE632 pKa = 4.72MPTEE636 pKa = 4.27DD637 pKa = 4.96APPLCKK643 pKa = 9.83PIEE646 pKa = 4.28ATPQDD651 pKa = 3.49ILII654 pKa = 4.57
MM1 pKa = 7.18NVGGFSEE8 pKa = 5.17SIWHH12 pKa = 5.33FAGYY16 pKa = 10.15LRR18 pKa = 11.84LIEE21 pKa = 4.14PAARR25 pKa = 11.84PSDD28 pKa = 4.41LFDD31 pKa = 5.24GDD33 pKa = 4.49PLPPPGDD40 pKa = 3.77EE41 pKa = 3.78IYY43 pKa = 11.25GRR45 pKa = 11.84LSEE48 pKa = 4.6TLPPAQFEE56 pKa = 4.41EE57 pKa = 4.75TASQRR62 pKa = 11.84VLFVEE67 pKa = 5.27APAASPALPGLKK79 pKa = 9.8LASPDD84 pKa = 3.79FKK86 pKa = 10.85PISLNPSSFPALPRR100 pKa = 11.84RR101 pKa = 11.84DD102 pKa = 4.13APLQEE107 pKa = 4.38PDD109 pKa = 2.98IALEE113 pKa = 3.98GGIQFRR119 pKa = 11.84FLSQSRR125 pKa = 11.84SISVDD130 pKa = 3.34YY131 pKa = 11.04QPGGTEE137 pKa = 3.6TLLQLRR143 pKa = 11.84QINTMDD149 pKa = 3.95DD150 pKa = 3.5RR151 pKa = 11.84DD152 pKa = 4.34VILSDD157 pKa = 4.0ALPGVLPPEE166 pKa = 4.61IEE168 pKa = 3.89TGAAVAEE175 pKa = 4.33MLQQAQEE182 pKa = 4.13ATPDD186 pKa = 3.67SLPFEE191 pKa = 5.57AIGSTQALIAAITARR206 pKa = 11.84DD207 pKa = 3.7AAWAQSGQSPHH218 pKa = 7.19ADD220 pKa = 3.15PDD222 pKa = 3.7YY223 pKa = 10.21TPPSGLIVDD232 pKa = 4.6GVVVAAAPTGPTIEE246 pKa = 4.99QIAPWRR252 pKa = 11.84EE253 pKa = 3.61AASAPTQFVSQTVSEE268 pKa = 4.26AASTLGIGAVAEE280 pKa = 4.35TGLNQQINQAVILDD294 pKa = 3.94LNEE297 pKa = 4.45AVGSMIVGGDD307 pKa = 3.15AFFSRR312 pKa = 11.84GIVQVNILTDD322 pKa = 3.55SDD324 pKa = 4.05HH325 pKa = 8.05VDD327 pKa = 3.07IAIEE331 pKa = 4.18GAGSVGVFTTDD342 pKa = 3.13NQLHH346 pKa = 6.54NIAEE350 pKa = 4.49FVTHH354 pKa = 7.46DD355 pKa = 3.08MTARR359 pKa = 11.84YY360 pKa = 9.24SGAAATPYY368 pKa = 9.49WHH370 pKa = 7.27VDD372 pKa = 3.11TLAGNFYY379 pKa = 10.6DD380 pKa = 4.91VKK382 pKa = 10.54TLVQFNGLEE391 pKa = 4.35DD392 pKa = 3.86GDD394 pKa = 3.87RR395 pKa = 11.84TVQASEE401 pKa = 3.84GAYY404 pKa = 10.36FEE406 pKa = 5.6AKK408 pKa = 9.51TGLNGQVNYY417 pKa = 11.45AMVTSLDD424 pKa = 3.4QYY426 pKa = 11.33DD427 pKa = 4.05IIIIAGDD434 pKa = 3.3YY435 pKa = 10.6HH436 pKa = 6.17RR437 pKa = 11.84ADD439 pKa = 3.93WIFQYY444 pKa = 11.34NIMLDD449 pKa = 3.39SDD451 pKa = 4.07VAKK454 pKa = 10.58LYY456 pKa = 10.78AAGGTAEE463 pKa = 5.39DD464 pKa = 4.59DD465 pKa = 3.99VAVTTGFNSLTNSASITTYY484 pKa = 11.21DD485 pKa = 3.26SAAFAALNAAQHH497 pKa = 6.65DD498 pKa = 4.75LLSALAQGATVLTPHH513 pKa = 7.29ADD515 pKa = 2.69WGLNGNISGEE525 pKa = 4.09LKK527 pKa = 10.15ILYY530 pKa = 10.18VSGDD534 pKa = 3.8YY535 pKa = 11.27YY536 pKa = 11.28DD537 pKa = 4.3VNVITQINLMVDD549 pKa = 3.52ADD551 pKa = 3.82QAIQASAIGGEE562 pKa = 4.26LGVAAGGNSAANYY575 pKa = 10.66AEE577 pKa = 4.81IIDD580 pKa = 4.79PGTLSTSRR588 pKa = 11.84YY589 pKa = 8.96IGGEE593 pKa = 4.0AYY595 pKa = 10.44DD596 pKa = 4.44DD597 pKa = 5.19AILVQTNLVTDD608 pKa = 4.54DD609 pKa = 4.06DD610 pKa = 4.68TVTIHH615 pKa = 7.14DD616 pKa = 4.08TTTLVPEE623 pKa = 4.75FVAFAEE629 pKa = 4.27QTEE632 pKa = 4.72MPTEE636 pKa = 4.27DD637 pKa = 4.96APPLCKK643 pKa = 9.83PIEE646 pKa = 4.28ATPQDD651 pKa = 3.49ILII654 pKa = 4.57
Molecular weight: 69.14 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q6KBN6|A0A0Q6KBN6_9BRAD GCN5 family acetyltransferase OS=Bosea sp. Leaf344 OX=1736346 GN=ASG72_07820 PE=4 SV=1
MM1 pKa = 7.55SISGSLVPVAVGAVAVVLLLGLINMLRR28 pKa = 11.84GGSPGTSQRR37 pKa = 11.84LMRR40 pKa = 11.84LRR42 pKa = 11.84VILQFIAILVILGVLWWRR60 pKa = 11.84SGG62 pKa = 3.16
MM1 pKa = 7.55SISGSLVPVAVGAVAVVLLLGLINMLRR28 pKa = 11.84GGSPGTSQRR37 pKa = 11.84LMRR40 pKa = 11.84LRR42 pKa = 11.84VILQFIAILVILGVLWWRR60 pKa = 11.84SGG62 pKa = 3.16
Molecular weight: 6.57 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1240282 |
41 |
3073 |
320.0 |
34.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.91 ± 0.054 | 0.82 ± 0.011 |
5.252 ± 0.035 | 5.577 ± 0.036 |
3.586 ± 0.023 | 9.018 ± 0.041 |
1.907 ± 0.018 | 5.057 ± 0.031 |
2.854 ± 0.031 | 10.777 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.355 ± 0.018 | 2.135 ± 0.024 |
5.486 ± 0.034 | 3.159 ± 0.019 |
7.581 ± 0.041 | 5.111 ± 0.029 |
4.932 ± 0.027 | 7.214 ± 0.035 |
1.273 ± 0.018 | 1.996 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
