
Zahedan rhabdovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Zarhavirus; Zahedan zarhavirus
Average proteome isoelectric point is 6.37
Get precalculated fractions of proteins
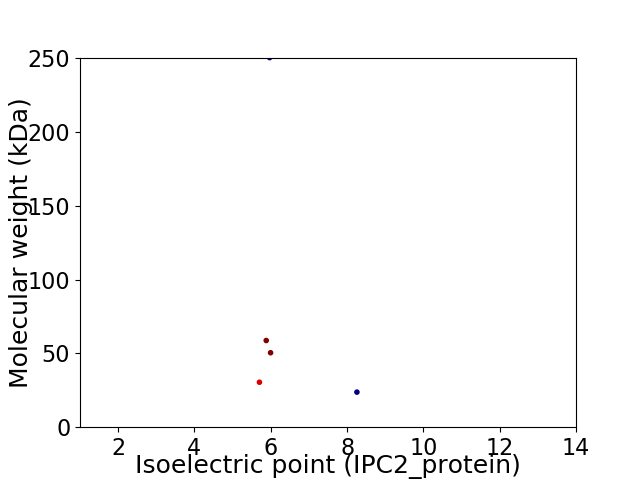
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
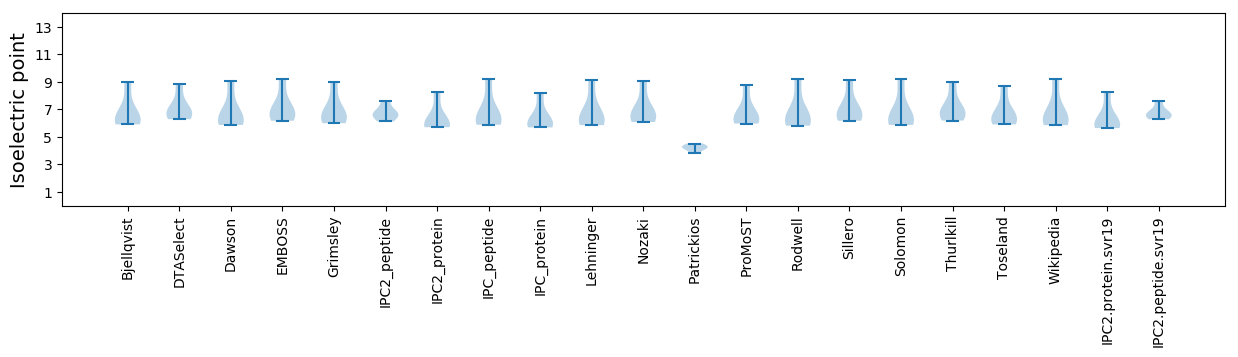
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0R6CHI1|A0A0R6CHI1_9RHAB Putative matrix protein OS=Zahedan rhabdovirus OX=1620507 GN=M PE=4 SV=1
MM1 pKa = 7.62SKK3 pKa = 10.34RR4 pKa = 11.84FRR6 pKa = 11.84VPSYY10 pKa = 10.29DD11 pKa = 3.39PSRR14 pKa = 11.84MVEE17 pKa = 3.99VAKK20 pKa = 10.72AADD23 pKa = 3.29NAYY26 pKa = 9.79EE27 pKa = 4.13NEE29 pKa = 4.0VEE31 pKa = 4.19KK32 pKa = 11.32DD33 pKa = 3.04KK34 pKa = 11.14DD35 pKa = 3.75IPPLTQALSAITLTRR50 pKa = 11.84EE51 pKa = 3.52EE52 pKa = 4.28EE53 pKa = 3.98ARR55 pKa = 11.84LGDD58 pKa = 4.32DD59 pKa = 4.59SYY61 pKa = 12.1LSDD64 pKa = 4.09SDD66 pKa = 5.09LDD68 pKa = 4.62RR69 pKa = 11.84SDD71 pKa = 5.07KK72 pKa = 11.28DD73 pKa = 4.0DD74 pKa = 5.25DD75 pKa = 4.18EE76 pKa = 4.82STPSSCSSHH85 pKa = 6.73RR86 pKa = 11.84FADD89 pKa = 3.76SEE91 pKa = 4.3GAKK94 pKa = 10.03AQFYY98 pKa = 10.84RR99 pKa = 11.84KK100 pKa = 9.42INFDD104 pKa = 3.37RR105 pKa = 11.84GIEE108 pKa = 3.85YY109 pKa = 9.81HH110 pKa = 6.85IPEE113 pKa = 5.69KK114 pKa = 10.24ITTPQDD120 pKa = 3.94AIDD123 pKa = 4.7DD124 pKa = 4.24LLSYY128 pKa = 9.83MEE130 pKa = 5.07DD131 pKa = 2.74MGMIDD136 pKa = 3.91SYY138 pKa = 11.48RR139 pKa = 11.84IDD141 pKa = 3.25FRR143 pKa = 11.84NRR145 pKa = 11.84RR146 pKa = 11.84KK147 pKa = 10.29VSINPRR153 pKa = 11.84STIPVRR159 pKa = 11.84DD160 pKa = 4.29EE161 pKa = 3.67IPEE164 pKa = 3.99EE165 pKa = 4.49RR166 pKa = 11.84FTPPPIKK173 pKa = 10.04MIVQDD178 pKa = 4.29PPKK181 pKa = 10.46ADD183 pKa = 3.04TGAIAKK189 pKa = 7.49STPKK193 pKa = 9.44STPKK197 pKa = 9.97KK198 pKa = 8.98VKK200 pKa = 9.72AKK202 pKa = 10.1KK203 pKa = 10.18VSLPSSGKK211 pKa = 10.19LPVYY215 pKa = 10.2QYY217 pKa = 9.54ITIYY221 pKa = 10.44GKK223 pKa = 10.79KK224 pKa = 10.13LDD226 pKa = 3.88IDD228 pKa = 4.21LNLYY232 pKa = 10.58CGGCDD237 pKa = 4.43LTDD240 pKa = 4.24FDD242 pKa = 6.31DD243 pKa = 3.93KK244 pKa = 11.52KK245 pKa = 10.21KK246 pKa = 9.58WYY248 pKa = 8.78YY249 pKa = 10.44HH250 pKa = 7.07AICSHH255 pKa = 6.77KK256 pKa = 10.46IKK258 pKa = 10.49GIKK261 pKa = 9.62LISIDD266 pKa = 3.73MNN268 pKa = 3.85
MM1 pKa = 7.62SKK3 pKa = 10.34RR4 pKa = 11.84FRR6 pKa = 11.84VPSYY10 pKa = 10.29DD11 pKa = 3.39PSRR14 pKa = 11.84MVEE17 pKa = 3.99VAKK20 pKa = 10.72AADD23 pKa = 3.29NAYY26 pKa = 9.79EE27 pKa = 4.13NEE29 pKa = 4.0VEE31 pKa = 4.19KK32 pKa = 11.32DD33 pKa = 3.04KK34 pKa = 11.14DD35 pKa = 3.75IPPLTQALSAITLTRR50 pKa = 11.84EE51 pKa = 3.52EE52 pKa = 4.28EE53 pKa = 3.98ARR55 pKa = 11.84LGDD58 pKa = 4.32DD59 pKa = 4.59SYY61 pKa = 12.1LSDD64 pKa = 4.09SDD66 pKa = 5.09LDD68 pKa = 4.62RR69 pKa = 11.84SDD71 pKa = 5.07KK72 pKa = 11.28DD73 pKa = 4.0DD74 pKa = 5.25DD75 pKa = 4.18EE76 pKa = 4.82STPSSCSSHH85 pKa = 6.73RR86 pKa = 11.84FADD89 pKa = 3.76SEE91 pKa = 4.3GAKK94 pKa = 10.03AQFYY98 pKa = 10.84RR99 pKa = 11.84KK100 pKa = 9.42INFDD104 pKa = 3.37RR105 pKa = 11.84GIEE108 pKa = 3.85YY109 pKa = 9.81HH110 pKa = 6.85IPEE113 pKa = 5.69KK114 pKa = 10.24ITTPQDD120 pKa = 3.94AIDD123 pKa = 4.7DD124 pKa = 4.24LLSYY128 pKa = 9.83MEE130 pKa = 5.07DD131 pKa = 2.74MGMIDD136 pKa = 3.91SYY138 pKa = 11.48RR139 pKa = 11.84IDD141 pKa = 3.25FRR143 pKa = 11.84NRR145 pKa = 11.84RR146 pKa = 11.84KK147 pKa = 10.29VSINPRR153 pKa = 11.84STIPVRR159 pKa = 11.84DD160 pKa = 4.29EE161 pKa = 3.67IPEE164 pKa = 3.99EE165 pKa = 4.49RR166 pKa = 11.84FTPPPIKK173 pKa = 10.04MIVQDD178 pKa = 4.29PPKK181 pKa = 10.46ADD183 pKa = 3.04TGAIAKK189 pKa = 7.49STPKK193 pKa = 9.44STPKK197 pKa = 9.97KK198 pKa = 8.98VKK200 pKa = 9.72AKK202 pKa = 10.1KK203 pKa = 10.18VSLPSSGKK211 pKa = 10.19LPVYY215 pKa = 10.2QYY217 pKa = 9.54ITIYY221 pKa = 10.44GKK223 pKa = 10.79KK224 pKa = 10.13LDD226 pKa = 3.88IDD228 pKa = 4.21LNLYY232 pKa = 10.58CGGCDD237 pKa = 4.43LTDD240 pKa = 4.24FDD242 pKa = 6.31DD243 pKa = 3.93KK244 pKa = 11.52KK245 pKa = 10.21KK246 pKa = 9.58WYY248 pKa = 8.78YY249 pKa = 10.44HH250 pKa = 7.07AICSHH255 pKa = 6.77KK256 pKa = 10.46IKK258 pKa = 10.49GIKK261 pKa = 9.62LISIDD266 pKa = 3.73MNN268 pKa = 3.85
Molecular weight: 30.47 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0R6CHI1|A0A0R6CHI1_9RHAB Putative matrix protein OS=Zahedan rhabdovirus OX=1620507 GN=M PE=4 SV=1
MM1 pKa = 7.55NVITWTKK8 pKa = 10.42GKK10 pKa = 10.32VSQAISKK17 pKa = 10.15RR18 pKa = 11.84GKK20 pKa = 10.41KK21 pKa = 9.78EE22 pKa = 4.21DD23 pKa = 3.84GDD25 pKa = 4.52DD26 pKa = 3.7GAKK29 pKa = 9.91IPSYY33 pKa = 10.94DD34 pKa = 4.12LTPSAPPDD42 pKa = 3.58DD43 pKa = 4.96GYY45 pKa = 11.24QYY47 pKa = 10.22RR48 pKa = 11.84TINVEE53 pKa = 3.82AEE55 pKa = 4.07VDD57 pKa = 3.7VSVCLDD63 pKa = 3.2KK64 pKa = 11.54FEE66 pKa = 4.67TRR68 pKa = 11.84MEE70 pKa = 4.86AIVAPLINLHH80 pKa = 6.09MDD82 pKa = 3.28YY83 pKa = 10.56KK84 pKa = 11.44GEE86 pKa = 3.87GRR88 pKa = 11.84YY89 pKa = 10.2KK90 pKa = 10.61NIWLPLVMVAVFGLKK105 pKa = 10.29LGQRR109 pKa = 11.84SRR111 pKa = 11.84DD112 pKa = 3.35NTVYY116 pKa = 10.57KK117 pKa = 10.67VQFGKK122 pKa = 10.73GLKK125 pKa = 10.23LKK127 pKa = 10.2IKK129 pKa = 9.69SRR131 pKa = 11.84RR132 pKa = 11.84PFEE135 pKa = 4.01MDD137 pKa = 4.88RR138 pKa = 11.84KK139 pKa = 9.97ISWNQFQKK147 pKa = 10.8YY148 pKa = 8.8LCNGLTSEE156 pKa = 4.03WSIRR160 pKa = 11.84CQLEE164 pKa = 4.07YY165 pKa = 10.96TLVDD169 pKa = 3.26YY170 pKa = 11.52SMKK173 pKa = 10.69HH174 pKa = 4.35VDD176 pKa = 5.19KK177 pKa = 11.1EE178 pKa = 3.87LLQYY182 pKa = 10.26LRR184 pKa = 11.84AMRR187 pKa = 11.84VRR189 pKa = 11.84FEE191 pKa = 3.86VDD193 pKa = 2.71EE194 pKa = 4.27NQNLVLIMDD203 pKa = 4.49YY204 pKa = 11.46
MM1 pKa = 7.55NVITWTKK8 pKa = 10.42GKK10 pKa = 10.32VSQAISKK17 pKa = 10.15RR18 pKa = 11.84GKK20 pKa = 10.41KK21 pKa = 9.78EE22 pKa = 4.21DD23 pKa = 3.84GDD25 pKa = 4.52DD26 pKa = 3.7GAKK29 pKa = 9.91IPSYY33 pKa = 10.94DD34 pKa = 4.12LTPSAPPDD42 pKa = 3.58DD43 pKa = 4.96GYY45 pKa = 11.24QYY47 pKa = 10.22RR48 pKa = 11.84TINVEE53 pKa = 3.82AEE55 pKa = 4.07VDD57 pKa = 3.7VSVCLDD63 pKa = 3.2KK64 pKa = 11.54FEE66 pKa = 4.67TRR68 pKa = 11.84MEE70 pKa = 4.86AIVAPLINLHH80 pKa = 6.09MDD82 pKa = 3.28YY83 pKa = 10.56KK84 pKa = 11.44GEE86 pKa = 3.87GRR88 pKa = 11.84YY89 pKa = 10.2KK90 pKa = 10.61NIWLPLVMVAVFGLKK105 pKa = 10.29LGQRR109 pKa = 11.84SRR111 pKa = 11.84DD112 pKa = 3.35NTVYY116 pKa = 10.57KK117 pKa = 10.67VQFGKK122 pKa = 10.73GLKK125 pKa = 10.23LKK127 pKa = 10.2IKK129 pKa = 9.69SRR131 pKa = 11.84RR132 pKa = 11.84PFEE135 pKa = 4.01MDD137 pKa = 4.88RR138 pKa = 11.84KK139 pKa = 9.97ISWNQFQKK147 pKa = 10.8YY148 pKa = 8.8LCNGLTSEE156 pKa = 4.03WSIRR160 pKa = 11.84CQLEE164 pKa = 4.07YY165 pKa = 10.96TLVDD169 pKa = 3.26YY170 pKa = 11.52SMKK173 pKa = 10.69HH174 pKa = 4.35VDD176 pKa = 5.19KK177 pKa = 11.1EE178 pKa = 3.87LLQYY182 pKa = 10.26LRR184 pKa = 11.84AMRR187 pKa = 11.84VRR189 pKa = 11.84FEE191 pKa = 3.86VDD193 pKa = 2.71EE194 pKa = 4.27NQNLVLIMDD203 pKa = 4.49YY204 pKa = 11.46
Molecular weight: 23.78 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3603 |
204 |
2170 |
720.6 |
82.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.079 ± 0.785 | 1.665 ± 0.446 |
6.384 ± 0.705 | 6.495 ± 0.236 |
4.191 ± 0.282 | 5.828 ± 0.28 |
2.609 ± 0.382 | 6.245 ± 0.282 |
5.773 ± 0.74 | 9.77 ± 0.987 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.526 ± 0.391 | 4.33 ± 0.314 |
4.746 ± 0.495 | 3.053 ± 0.324 |
6.328 ± 0.349 | 6.273 ± 0.578 |
6.245 ± 0.332 | 6.273 ± 0.433 |
1.971 ± 0.341 | 4.219 ± 0.203 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
