
Cercopithecine alphaherpesvirus 2
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Herpesviridae; Alphaherpesvirinae; Simplexvirus
Average proteome isoelectric point is 7.14
Get precalculated fractions of proteins
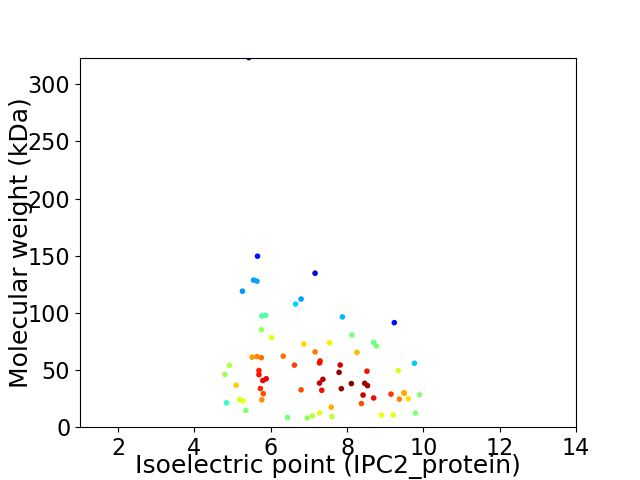
Virtual 2D-PAGE plot for 73 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
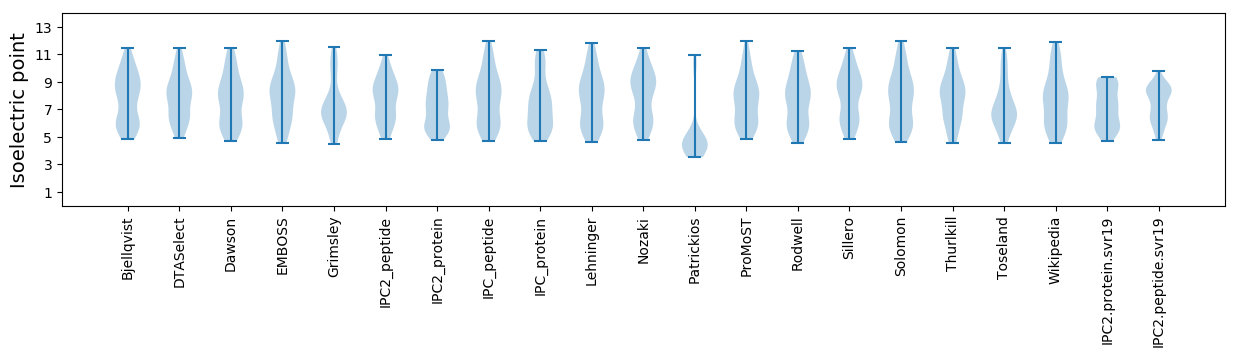
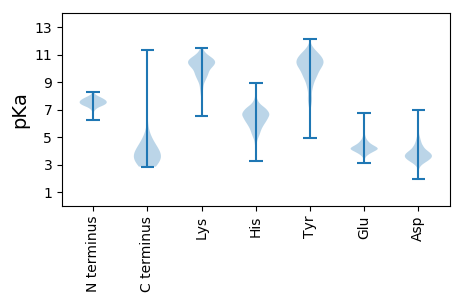
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q5Y0V0|Q5Y0V0_9ALPH UL3 OS=Cercopithecine alphaherpesvirus 2 OX=10317 GN=UL3 PE=3 SV=1
MM1 pKa = 7.22SASASSATIAYY12 pKa = 8.63SLHH15 pKa = 5.89GAATSSACVLPDD27 pKa = 3.52AEE29 pKa = 4.17QVVCAFEE36 pKa = 4.29SGTRR40 pKa = 11.84AIASRR45 pKa = 11.84GCLRR49 pKa = 11.84HH50 pKa = 6.6DD51 pKa = 3.74ALSRR55 pKa = 11.84GAVVVRR61 pKa = 11.84QTPVGLLVMVDD72 pKa = 4.01CRR74 pKa = 11.84TEE76 pKa = 3.92FCAYY80 pKa = 9.89RR81 pKa = 11.84FVGRR85 pKa = 11.84SEE87 pKa = 3.96RR88 pKa = 11.84QRR90 pKa = 11.84LEE92 pKa = 4.0RR93 pKa = 11.84WWDD96 pKa = 3.52TTLCAYY102 pKa = 9.69PFDD105 pKa = 3.98SWVSSTRR112 pKa = 11.84GEE114 pKa = 4.3SARR117 pKa = 11.84SPTAGIATVVWGEE130 pKa = 3.7DD131 pKa = 3.15SIYY134 pKa = 9.94ITVTVYY140 pKa = 10.46GSPPEE145 pKa = 4.56GEE147 pKa = 4.34AGAPPRR153 pKa = 11.84LPPPPGDD160 pKa = 4.34APDD163 pKa = 4.36APSPPAVSPASAEE176 pKa = 3.93TGAAADD182 pKa = 4.17LLVEE186 pKa = 4.19VMKK189 pKa = 10.32EE190 pKa = 3.84IQLSPTLGFGPCDD203 pKa = 3.21AA204 pKa = 5.68
MM1 pKa = 7.22SASASSATIAYY12 pKa = 8.63SLHH15 pKa = 5.89GAATSSACVLPDD27 pKa = 3.52AEE29 pKa = 4.17QVVCAFEE36 pKa = 4.29SGTRR40 pKa = 11.84AIASRR45 pKa = 11.84GCLRR49 pKa = 11.84HH50 pKa = 6.6DD51 pKa = 3.74ALSRR55 pKa = 11.84GAVVVRR61 pKa = 11.84QTPVGLLVMVDD72 pKa = 4.01CRR74 pKa = 11.84TEE76 pKa = 3.92FCAYY80 pKa = 9.89RR81 pKa = 11.84FVGRR85 pKa = 11.84SEE87 pKa = 3.96RR88 pKa = 11.84QRR90 pKa = 11.84LEE92 pKa = 4.0RR93 pKa = 11.84WWDD96 pKa = 3.52TTLCAYY102 pKa = 9.69PFDD105 pKa = 3.98SWVSSTRR112 pKa = 11.84GEE114 pKa = 4.3SARR117 pKa = 11.84SPTAGIATVVWGEE130 pKa = 3.7DD131 pKa = 3.15SIYY134 pKa = 9.94ITVTVYY140 pKa = 10.46GSPPEE145 pKa = 4.56GEE147 pKa = 4.34AGAPPRR153 pKa = 11.84LPPPPGDD160 pKa = 4.34APDD163 pKa = 4.36APSPPAVSPASAEE176 pKa = 3.93TGAAADD182 pKa = 4.17LLVEE186 pKa = 4.19VMKK189 pKa = 10.32EE190 pKa = 3.84IQLSPTLGFGPCDD203 pKa = 3.21AA204 pKa = 5.68
Molecular weight: 21.32 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q5Y0V1|Q5Y0V1_9ALPH Uracil-DNA glycosylase OS=Cercopithecine alphaherpesvirus 2 OX=10317 GN=UL2 PE=3 SV=1
MM1 pKa = 7.16SAVGAGAVPSTLAVLASWGWAFAPHH26 pKa = 7.31DD27 pKa = 4.55GPPARR32 pKa = 11.84AAEE35 pKa = 4.6APASPAPEE43 pKa = 4.6APTGHH48 pKa = 7.13PVPPRR53 pKa = 11.84ADD55 pKa = 3.28DD56 pKa = 3.68RR57 pKa = 11.84APPGVRR63 pKa = 11.84APDD66 pKa = 4.15PDD68 pKa = 3.73HH69 pKa = 6.93VAFDD73 pKa = 3.73TMFMVSSVDD82 pKa = 3.32EE83 pKa = 4.19LGRR86 pKa = 11.84RR87 pKa = 11.84QLTDD91 pKa = 3.82TIRR94 pKa = 11.84KK95 pKa = 9.09DD96 pKa = 3.43LRR98 pKa = 11.84LSLVNLAIACTKK110 pKa = 9.72TSSFSGTAGRR120 pKa = 11.84ARR122 pKa = 11.84PRR124 pKa = 11.84ARR126 pKa = 11.84HH127 pKa = 5.4GPGHH131 pKa = 6.83RR132 pKa = 11.84EE133 pKa = 4.0PPSHH137 pKa = 7.31KK138 pKa = 9.61SLQMFVLCQRR148 pKa = 11.84PDD150 pKa = 2.99ATRR153 pKa = 11.84VRR155 pKa = 11.84DD156 pKa = 3.44QLRR159 pKa = 11.84AVIASRR165 pKa = 11.84KK166 pKa = 6.69PRR168 pKa = 11.84KK169 pKa = 9.7YY170 pKa = 8.92YY171 pKa = 9.88TRR173 pKa = 11.84SSDD176 pKa = 3.26GRR178 pKa = 11.84ISPAVPVFVHH188 pKa = 6.3EE189 pKa = 4.83FVSDD193 pKa = 3.9APVHH197 pKa = 4.4VHH199 pKa = 6.69RR200 pKa = 11.84DD201 pKa = 3.43NVIAPVVPRR210 pKa = 11.84RR211 pKa = 11.84RR212 pKa = 11.84TSTRR216 pKa = 11.84LADD219 pKa = 4.21PASEE223 pKa = 5.39DD224 pKa = 3.13PWQQ227 pKa = 3.83
MM1 pKa = 7.16SAVGAGAVPSTLAVLASWGWAFAPHH26 pKa = 7.31DD27 pKa = 4.55GPPARR32 pKa = 11.84AAEE35 pKa = 4.6APASPAPEE43 pKa = 4.6APTGHH48 pKa = 7.13PVPPRR53 pKa = 11.84ADD55 pKa = 3.28DD56 pKa = 3.68RR57 pKa = 11.84APPGVRR63 pKa = 11.84APDD66 pKa = 4.15PDD68 pKa = 3.73HH69 pKa = 6.93VAFDD73 pKa = 3.73TMFMVSSVDD82 pKa = 3.32EE83 pKa = 4.19LGRR86 pKa = 11.84RR87 pKa = 11.84QLTDD91 pKa = 3.82TIRR94 pKa = 11.84KK95 pKa = 9.09DD96 pKa = 3.43LRR98 pKa = 11.84LSLVNLAIACTKK110 pKa = 9.72TSSFSGTAGRR120 pKa = 11.84ARR122 pKa = 11.84PRR124 pKa = 11.84ARR126 pKa = 11.84HH127 pKa = 5.4GPGHH131 pKa = 6.83RR132 pKa = 11.84EE133 pKa = 4.0PPSHH137 pKa = 7.31KK138 pKa = 9.61SLQMFVLCQRR148 pKa = 11.84PDD150 pKa = 2.99ATRR153 pKa = 11.84VRR155 pKa = 11.84DD156 pKa = 3.44QLRR159 pKa = 11.84AVIASRR165 pKa = 11.84KK166 pKa = 6.69PRR168 pKa = 11.84KK169 pKa = 9.7YY170 pKa = 8.92YY171 pKa = 9.88TRR173 pKa = 11.84SSDD176 pKa = 3.26GRR178 pKa = 11.84ISPAVPVFVHH188 pKa = 6.3EE189 pKa = 4.83FVSDD193 pKa = 3.9APVHH197 pKa = 4.4VHH199 pKa = 6.69RR200 pKa = 11.84DD201 pKa = 3.43NVIAPVVPRR210 pKa = 11.84RR211 pKa = 11.84RR212 pKa = 11.84TSTRR216 pKa = 11.84LADD219 pKa = 4.21PASEE223 pKa = 5.39DD224 pKa = 3.13PWQQ227 pKa = 3.83
Molecular weight: 24.57 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
37766 |
77 |
3070 |
517.3 |
55.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
15.946 ± 0.576 | 1.729 ± 0.099 |
5.33 ± 0.168 | 5.452 ± 0.167 |
3.344 ± 0.186 | 8.092 ± 0.18 |
2.29 ± 0.119 | 2.063 ± 0.153 |
1.157 ± 0.124 | 9.956 ± 0.229 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.395 ± 0.107 | 1.673 ± 0.16 |
9.286 ± 0.462 | 2.261 ± 0.131 |
9.744 ± 0.215 | 5.108 ± 0.18 |
4.753 ± 0.153 | 6.919 ± 0.229 |
1.123 ± 0.066 | 2.378 ± 0.136 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
