
Lolium perenne-associated virus
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses
Average proteome isoelectric point is 8.06
Get precalculated fractions of proteins
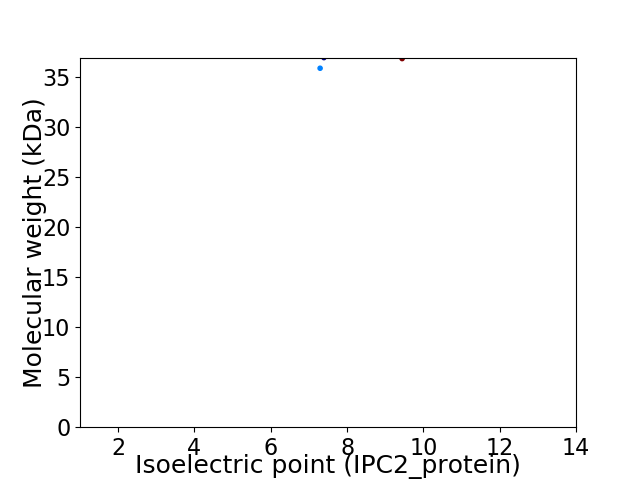
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A345BKA8|A0A345BKA8_9VIRU Replication-associated protein OS=Lolium perenne-associated virus OX=2282644 PE=3 SV=1
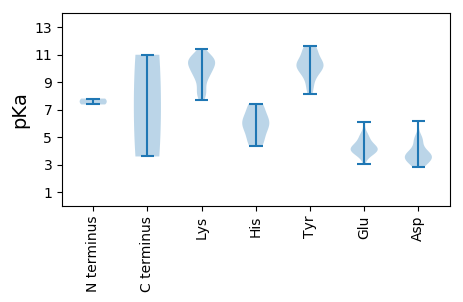
MM1 pKa = 7.8SFRR4 pKa = 11.84LQSKK8 pKa = 9.94ILLLTYY14 pKa = 10.34SQADD18 pKa = 4.01AIDD21 pKa = 3.55HH22 pKa = 5.82EE23 pKa = 4.78RR24 pKa = 11.84LFKK27 pKa = 10.6FLSGLKK33 pKa = 9.09QPIRR37 pKa = 11.84IRR39 pKa = 11.84VGKK42 pKa = 8.18EE43 pKa = 3.04AHH45 pKa = 6.34MDD47 pKa = 3.35GHH49 pKa = 5.28THH51 pKa = 4.32YY52 pKa = 10.3HH53 pKa = 7.35AYY55 pKa = 10.46AEE57 pKa = 4.25WEE59 pKa = 4.38KK60 pKa = 11.03KK61 pKa = 10.6YY62 pKa = 9.18EE63 pKa = 4.25TTDD66 pKa = 2.89ARR68 pKa = 11.84HH69 pKa = 6.12FDD71 pKa = 3.86FEE73 pKa = 5.21SIHH76 pKa = 6.25PNIVSRR82 pKa = 11.84VRR84 pKa = 11.84DD85 pKa = 3.69PLSAWNYY92 pKa = 8.56CGKK95 pKa = 10.43DD96 pKa = 3.46GVAIDD101 pKa = 4.43YY102 pKa = 9.85GEE104 pKa = 4.62CPQPRR109 pKa = 11.84AINKK113 pKa = 8.4RR114 pKa = 11.84KK115 pKa = 10.21QMMDD119 pKa = 2.85AFDD122 pKa = 3.72ASSRR126 pKa = 11.84QEE128 pKa = 3.78FMEE131 pKa = 4.5MIKK134 pKa = 10.7NADD137 pKa = 3.34PGRR140 pKa = 11.84YY141 pKa = 8.65IFQHH145 pKa = 5.53QNLEE149 pKa = 4.13YY150 pKa = 10.21FANKK154 pKa = 8.88QFAVCIPQYY163 pKa = 11.24VSDD166 pKa = 5.08PDD168 pKa = 4.51LIFNIPDD175 pKa = 3.68EE176 pKa = 4.37LTTWLSQRR184 pKa = 11.84HH185 pKa = 4.72EE186 pKa = 4.04ADD188 pKa = 3.56RR189 pKa = 11.84PKK191 pKa = 11.04SLILTGPSRR200 pKa = 11.84TGKK203 pKa = 8.23TKK205 pKa = 9.29WARR208 pKa = 11.84SLGRR212 pKa = 11.84HH213 pKa = 6.08IYY215 pKa = 9.53WNGAFDD221 pKa = 6.19LSIFDD226 pKa = 4.8NDD228 pKa = 3.04AQYY231 pKa = 11.39AIFDD235 pKa = 5.0DD236 pKa = 4.82FPDD239 pKa = 3.04WDD241 pKa = 3.84RR242 pKa = 11.84FVQYY246 pKa = 10.15KK247 pKa = 8.91QWLGAQQHH255 pKa = 5.3FTATDD260 pKa = 3.1KK261 pKa = 11.43YY262 pKa = 10.17MKK264 pKa = 9.23KK265 pKa = 7.79TQIKK269 pKa = 8.2WGKK272 pKa = 8.38PCIILSNQNPLFKK285 pKa = 10.42DD286 pKa = 3.54YY287 pKa = 11.45LWIKK291 pKa = 10.16EE292 pKa = 4.01NTIEE296 pKa = 5.2CEE298 pKa = 4.1IKK300 pKa = 9.78TKK302 pKa = 10.76LYY304 pKa = 10.98
MM1 pKa = 7.8SFRR4 pKa = 11.84LQSKK8 pKa = 9.94ILLLTYY14 pKa = 10.34SQADD18 pKa = 4.01AIDD21 pKa = 3.55HH22 pKa = 5.82EE23 pKa = 4.78RR24 pKa = 11.84LFKK27 pKa = 10.6FLSGLKK33 pKa = 9.09QPIRR37 pKa = 11.84IRR39 pKa = 11.84VGKK42 pKa = 8.18EE43 pKa = 3.04AHH45 pKa = 6.34MDD47 pKa = 3.35GHH49 pKa = 5.28THH51 pKa = 4.32YY52 pKa = 10.3HH53 pKa = 7.35AYY55 pKa = 10.46AEE57 pKa = 4.25WEE59 pKa = 4.38KK60 pKa = 11.03KK61 pKa = 10.6YY62 pKa = 9.18EE63 pKa = 4.25TTDD66 pKa = 2.89ARR68 pKa = 11.84HH69 pKa = 6.12FDD71 pKa = 3.86FEE73 pKa = 5.21SIHH76 pKa = 6.25PNIVSRR82 pKa = 11.84VRR84 pKa = 11.84DD85 pKa = 3.69PLSAWNYY92 pKa = 8.56CGKK95 pKa = 10.43DD96 pKa = 3.46GVAIDD101 pKa = 4.43YY102 pKa = 9.85GEE104 pKa = 4.62CPQPRR109 pKa = 11.84AINKK113 pKa = 8.4RR114 pKa = 11.84KK115 pKa = 10.21QMMDD119 pKa = 2.85AFDD122 pKa = 3.72ASSRR126 pKa = 11.84QEE128 pKa = 3.78FMEE131 pKa = 4.5MIKK134 pKa = 10.7NADD137 pKa = 3.34PGRR140 pKa = 11.84YY141 pKa = 8.65IFQHH145 pKa = 5.53QNLEE149 pKa = 4.13YY150 pKa = 10.21FANKK154 pKa = 8.88QFAVCIPQYY163 pKa = 11.24VSDD166 pKa = 5.08PDD168 pKa = 4.51LIFNIPDD175 pKa = 3.68EE176 pKa = 4.37LTTWLSQRR184 pKa = 11.84HH185 pKa = 4.72EE186 pKa = 4.04ADD188 pKa = 3.56RR189 pKa = 11.84PKK191 pKa = 11.04SLILTGPSRR200 pKa = 11.84TGKK203 pKa = 8.23TKK205 pKa = 9.29WARR208 pKa = 11.84SLGRR212 pKa = 11.84HH213 pKa = 6.08IYY215 pKa = 9.53WNGAFDD221 pKa = 6.19LSIFDD226 pKa = 4.8NDD228 pKa = 3.04AQYY231 pKa = 11.39AIFDD235 pKa = 5.0DD236 pKa = 4.82FPDD239 pKa = 3.04WDD241 pKa = 3.84RR242 pKa = 11.84FVQYY246 pKa = 10.15KK247 pKa = 8.91QWLGAQQHH255 pKa = 5.3FTATDD260 pKa = 3.1KK261 pKa = 11.43YY262 pKa = 10.17MKK264 pKa = 9.23KK265 pKa = 7.79TQIKK269 pKa = 8.2WGKK272 pKa = 8.38PCIILSNQNPLFKK285 pKa = 10.42DD286 pKa = 3.54YY287 pKa = 11.45LWIKK291 pKa = 10.16EE292 pKa = 4.01NTIEE296 pKa = 5.2CEE298 pKa = 4.1IKK300 pKa = 9.78TKK302 pKa = 10.76LYY304 pKa = 10.98
Molecular weight: 35.91 kDa
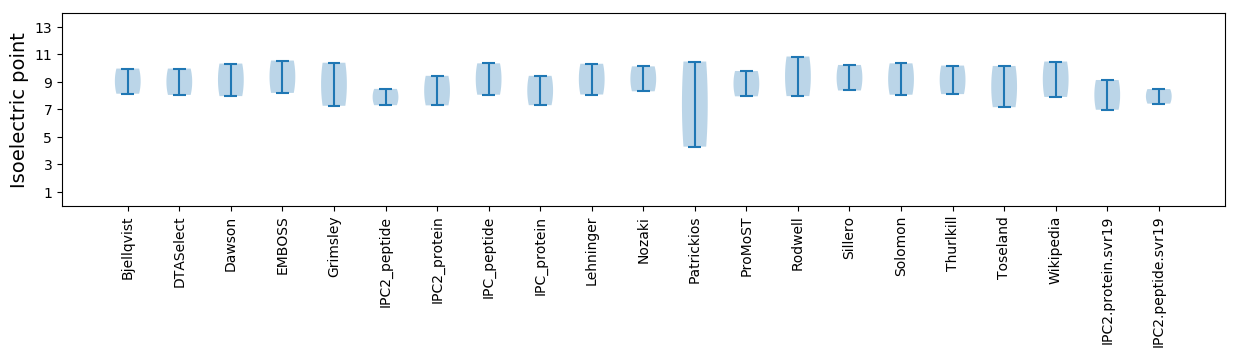
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A345BKA8|A0A345BKA8_9VIRU Replication-associated protein OS=Lolium perenne-associated virus OX=2282644 PE=3 SV=1
MM1 pKa = 7.37VRR3 pKa = 11.84GTRR6 pKa = 11.84KK7 pKa = 9.5RR8 pKa = 11.84KK9 pKa = 9.56FKK11 pKa = 10.28GGHH14 pKa = 4.58IAIPRR19 pKa = 11.84KK20 pKa = 9.43KK21 pKa = 10.5VKK23 pKa = 9.61TGFRR27 pKa = 11.84NIKK30 pKa = 9.67KK31 pKa = 10.1KK32 pKa = 10.03VGGRR36 pKa = 11.84RR37 pKa = 11.84GSSSGVVMQSGMTSKK52 pKa = 9.8TKK54 pKa = 10.73LLVAGHH60 pKa = 6.74KK61 pKa = 10.84LNTKK65 pKa = 7.69VLKK68 pKa = 10.62LLSNPIRR75 pKa = 11.84VIVEE79 pKa = 3.99GNKK82 pKa = 10.13SVGANGMNYY91 pKa = 10.63AKK93 pKa = 10.68GEE95 pKa = 4.22YY96 pKa = 9.99LYY98 pKa = 10.27MDD100 pKa = 4.76ASQFDD105 pKa = 3.48IMAQLGFRR113 pKa = 11.84QRR115 pKa = 11.84AVSASGSLPAAIPGRR130 pKa = 11.84TFKK133 pKa = 10.93AWIQSFYY140 pKa = 11.52VEE142 pKa = 3.7QKK144 pKa = 9.66YY145 pKa = 11.11INMSNMAVKK154 pKa = 10.5LIIYY158 pKa = 8.16DD159 pKa = 3.74VKK161 pKa = 11.04VKK163 pKa = 10.46RR164 pKa = 11.84DD165 pKa = 3.49TSSGPFTCVQNDD177 pKa = 3.58VAILNQNRR185 pKa = 11.84DD186 pKa = 3.35TTITNDD192 pKa = 3.16AALATLTPGYY202 pKa = 10.78SPMDD206 pKa = 3.46SLQFRR211 pKa = 11.84THH213 pKa = 4.48WKK215 pKa = 8.1IVRR218 pKa = 11.84RR219 pKa = 11.84VQVHH223 pKa = 6.2LDD225 pKa = 2.93AGGVHH230 pKa = 5.77EE231 pKa = 6.11HH232 pKa = 7.39IIQANYY238 pKa = 10.41NKK240 pKa = 10.07IISRR244 pKa = 11.84NVSVEE249 pKa = 3.67DD250 pKa = 4.03NYY252 pKa = 11.59LKK254 pKa = 10.64GYY256 pKa = 10.07SFGVYY261 pKa = 9.5IEE263 pKa = 4.15VQPYY267 pKa = 9.59PIVDD271 pKa = 4.04GVTHH275 pKa = 7.4ALPSVAPPASEE286 pKa = 4.05VLMTSVGKK294 pKa = 10.28FSFRR298 pKa = 11.84EE299 pKa = 3.68MGMTPEE305 pKa = 5.07HH306 pKa = 6.98IEE308 pKa = 3.88VTNIWTLATNPTTVVDD324 pKa = 4.08EE325 pKa = 4.83SGLVTPGVTVV335 pKa = 3.59
MM1 pKa = 7.37VRR3 pKa = 11.84GTRR6 pKa = 11.84KK7 pKa = 9.5RR8 pKa = 11.84KK9 pKa = 9.56FKK11 pKa = 10.28GGHH14 pKa = 4.58IAIPRR19 pKa = 11.84KK20 pKa = 9.43KK21 pKa = 10.5VKK23 pKa = 9.61TGFRR27 pKa = 11.84NIKK30 pKa = 9.67KK31 pKa = 10.1KK32 pKa = 10.03VGGRR36 pKa = 11.84RR37 pKa = 11.84GSSSGVVMQSGMTSKK52 pKa = 9.8TKK54 pKa = 10.73LLVAGHH60 pKa = 6.74KK61 pKa = 10.84LNTKK65 pKa = 7.69VLKK68 pKa = 10.62LLSNPIRR75 pKa = 11.84VIVEE79 pKa = 3.99GNKK82 pKa = 10.13SVGANGMNYY91 pKa = 10.63AKK93 pKa = 10.68GEE95 pKa = 4.22YY96 pKa = 9.99LYY98 pKa = 10.27MDD100 pKa = 4.76ASQFDD105 pKa = 3.48IMAQLGFRR113 pKa = 11.84QRR115 pKa = 11.84AVSASGSLPAAIPGRR130 pKa = 11.84TFKK133 pKa = 10.93AWIQSFYY140 pKa = 11.52VEE142 pKa = 3.7QKK144 pKa = 9.66YY145 pKa = 11.11INMSNMAVKK154 pKa = 10.5LIIYY158 pKa = 8.16DD159 pKa = 3.74VKK161 pKa = 11.04VKK163 pKa = 10.46RR164 pKa = 11.84DD165 pKa = 3.49TSSGPFTCVQNDD177 pKa = 3.58VAILNQNRR185 pKa = 11.84DD186 pKa = 3.35TTITNDD192 pKa = 3.16AALATLTPGYY202 pKa = 10.78SPMDD206 pKa = 3.46SLQFRR211 pKa = 11.84THH213 pKa = 4.48WKK215 pKa = 8.1IVRR218 pKa = 11.84RR219 pKa = 11.84VQVHH223 pKa = 6.2LDD225 pKa = 2.93AGGVHH230 pKa = 5.77EE231 pKa = 6.11HH232 pKa = 7.39IIQANYY238 pKa = 10.41NKK240 pKa = 10.07IISRR244 pKa = 11.84NVSVEE249 pKa = 3.67DD250 pKa = 4.03NYY252 pKa = 11.59LKK254 pKa = 10.64GYY256 pKa = 10.07SFGVYY261 pKa = 9.5IEE263 pKa = 4.15VQPYY267 pKa = 9.59PIVDD271 pKa = 4.04GVTHH275 pKa = 7.4ALPSVAPPASEE286 pKa = 4.05VLMTSVGKK294 pKa = 10.28FSFRR298 pKa = 11.84EE299 pKa = 3.68MGMTPEE305 pKa = 5.07HH306 pKa = 6.98IEE308 pKa = 3.88VTNIWTLATNPTTVVDD324 pKa = 4.08EE325 pKa = 4.83SGLVTPGVTVV335 pKa = 3.59
Molecular weight: 36.88 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
639 |
304 |
335 |
319.5 |
36.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.729 ± 0.123 | 0.939 ± 0.484 |
5.634 ± 1.551 | 4.069 ± 0.593 |
4.538 ± 0.948 | 6.416 ± 1.468 |
2.973 ± 0.442 | 7.042 ± 0.359 |
7.668 ± 0.155 | 6.416 ± 0.337 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.973 ± 0.46 | 4.695 ± 0.513 |
4.538 ± 0.046 | 4.695 ± 0.841 |
5.477 ± 0.079 | 6.416 ± 0.791 |
5.947 ± 0.92 | 6.729 ± 3.036 |
1.878 ± 0.742 | 4.225 ± 0.486 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
