
Alphaproteobacteria phage PhiJL001
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; Yuavirus; Alphaproteobacteria virus phiJl001
Average proteome isoelectric point is 6.32
Get precalculated fractions of proteins
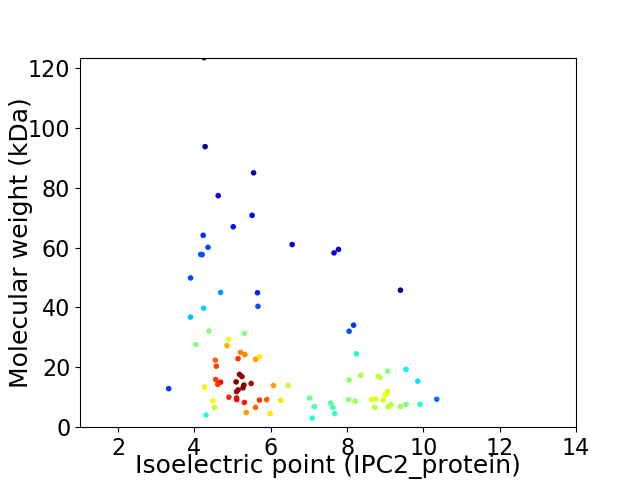
Virtual 2D-PAGE plot for 90 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
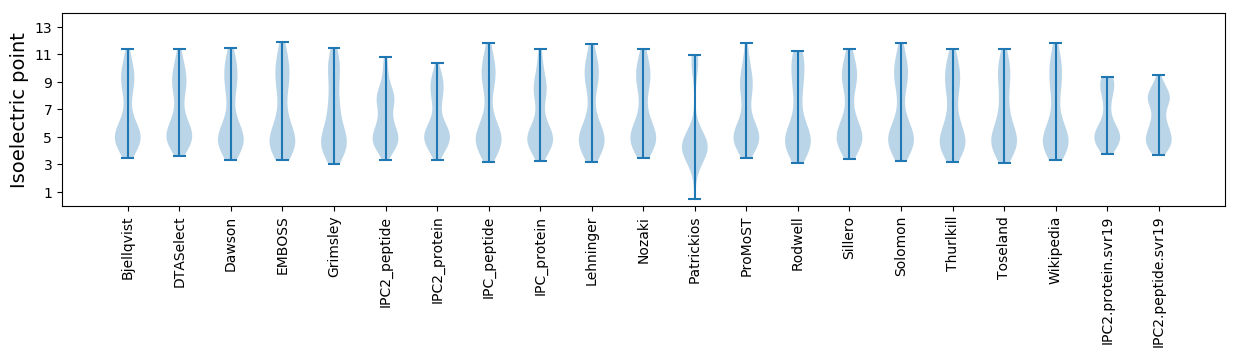
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q5DN34|Q5DN34_9CAUD Gp71 OS=Alphaproteobacteria phage PhiJL001 OX=2681607 GN=JL001p71 PE=4 SV=1
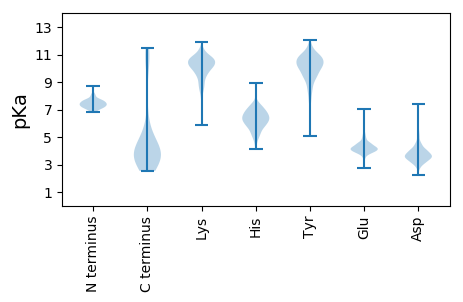
MM1 pKa = 7.48SLGTAYY7 pKa = 10.3GGSTTSSGGGVDD19 pKa = 3.66TTVEE23 pKa = 4.22TANSSGYY30 pKa = 9.68MYY32 pKa = 10.71LQGDD36 pKa = 4.06ATTEE40 pKa = 3.88GSVRR44 pKa = 11.84FNASSDD50 pKa = 2.72IGAGVFEE57 pKa = 4.5KK58 pKa = 10.92LEE60 pKa = 4.18NGIWTANTLEE70 pKa = 4.32MGVDD74 pKa = 3.73SLFLGPRR81 pKa = 11.84TGLASIGEE89 pKa = 4.27YY90 pKa = 10.76LSVEE94 pKa = 4.02NSNIPDD100 pKa = 3.36ARR102 pKa = 11.84RR103 pKa = 11.84LFPYY107 pKa = 9.8TVFNGTTIDD116 pKa = 3.99DD117 pKa = 4.57LATIPYY123 pKa = 7.9VTAHH127 pKa = 6.48LTNVVSVPFDD137 pKa = 3.63SDD139 pKa = 3.42EE140 pKa = 4.42TTGTSYY146 pKa = 11.02SWTFTGTVNAMLKK159 pKa = 10.4NIYY162 pKa = 9.89IKK164 pKa = 10.21IGSVAPTDD172 pKa = 3.5TVRR175 pKa = 11.84FFVRR179 pKa = 11.84DD180 pKa = 3.34NDD182 pKa = 3.73EE183 pKa = 4.09NGQIIFDD190 pKa = 3.75EE191 pKa = 4.88SYY193 pKa = 9.82PASAFAANSTVALQTTGFFHH213 pKa = 6.93NKK215 pKa = 9.74VGDD218 pKa = 3.77TYY220 pKa = 11.46YY221 pKa = 10.28STFEE225 pKa = 3.89SSQPFSILGSAALQLPYY242 pKa = 10.66FEE244 pKa = 5.17VDD246 pKa = 2.61ASFIRR251 pKa = 11.84DD252 pKa = 3.35DD253 pKa = 4.6RR254 pKa = 11.84LWQASPWEE262 pKa = 4.31AGTYY266 pKa = 10.42AVGDD270 pKa = 3.7YY271 pKa = 10.48VIEE274 pKa = 4.16GRR276 pKa = 11.84VPYY279 pKa = 10.05ICVVAGAQATDD290 pKa = 3.88FATNSTSWKK299 pKa = 9.13PINGYY304 pKa = 10.31DD305 pKa = 3.72FDD307 pKa = 4.99DD308 pKa = 3.46WVTNYY313 pKa = 7.48GTRR316 pKa = 11.84AILDD320 pKa = 3.68EE321 pKa = 4.75NYY323 pKa = 11.2DD324 pKa = 4.23LIIDD328 pKa = 4.08EE329 pKa = 4.66ATLEE333 pKa = 4.14PVVLEE338 pKa = 4.47GTT340 pKa = 3.79
MM1 pKa = 7.48SLGTAYY7 pKa = 10.3GGSTTSSGGGVDD19 pKa = 3.66TTVEE23 pKa = 4.22TANSSGYY30 pKa = 9.68MYY32 pKa = 10.71LQGDD36 pKa = 4.06ATTEE40 pKa = 3.88GSVRR44 pKa = 11.84FNASSDD50 pKa = 2.72IGAGVFEE57 pKa = 4.5KK58 pKa = 10.92LEE60 pKa = 4.18NGIWTANTLEE70 pKa = 4.32MGVDD74 pKa = 3.73SLFLGPRR81 pKa = 11.84TGLASIGEE89 pKa = 4.27YY90 pKa = 10.76LSVEE94 pKa = 4.02NSNIPDD100 pKa = 3.36ARR102 pKa = 11.84RR103 pKa = 11.84LFPYY107 pKa = 9.8TVFNGTTIDD116 pKa = 3.99DD117 pKa = 4.57LATIPYY123 pKa = 7.9VTAHH127 pKa = 6.48LTNVVSVPFDD137 pKa = 3.63SDD139 pKa = 3.42EE140 pKa = 4.42TTGTSYY146 pKa = 11.02SWTFTGTVNAMLKK159 pKa = 10.4NIYY162 pKa = 9.89IKK164 pKa = 10.21IGSVAPTDD172 pKa = 3.5TVRR175 pKa = 11.84FFVRR179 pKa = 11.84DD180 pKa = 3.34NDD182 pKa = 3.73EE183 pKa = 4.09NGQIIFDD190 pKa = 3.75EE191 pKa = 4.88SYY193 pKa = 9.82PASAFAANSTVALQTTGFFHH213 pKa = 6.93NKK215 pKa = 9.74VGDD218 pKa = 3.77TYY220 pKa = 11.46YY221 pKa = 10.28STFEE225 pKa = 3.89SSQPFSILGSAALQLPYY242 pKa = 10.66FEE244 pKa = 5.17VDD246 pKa = 2.61ASFIRR251 pKa = 11.84DD252 pKa = 3.35DD253 pKa = 4.6RR254 pKa = 11.84LWQASPWEE262 pKa = 4.31AGTYY266 pKa = 10.42AVGDD270 pKa = 3.7YY271 pKa = 10.48VIEE274 pKa = 4.16GRR276 pKa = 11.84VPYY279 pKa = 10.05ICVVAGAQATDD290 pKa = 3.88FATNSTSWKK299 pKa = 9.13PINGYY304 pKa = 10.31DD305 pKa = 3.72FDD307 pKa = 4.99DD308 pKa = 3.46WVTNYY313 pKa = 7.48GTRR316 pKa = 11.84AILDD320 pKa = 3.68EE321 pKa = 4.75NYY323 pKa = 11.2DD324 pKa = 4.23LIIDD328 pKa = 4.08EE329 pKa = 4.66ATLEE333 pKa = 4.14PVVLEE338 pKa = 4.47GTT340 pKa = 3.79
Molecular weight: 36.78 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q5DN44|Q5DN44_9CAUD Gp61 OS=Alphaproteobacteria phage PhiJL001 OX=2681607 GN=JL001p61 PE=4 SV=1
MM1 pKa = 7.37PTDD4 pKa = 3.67DD5 pKa = 5.34QIPGMPDD12 pKa = 2.81NVNEE16 pKa = 4.05EE17 pKa = 4.22WLDD20 pKa = 3.31AMIRR24 pKa = 11.84HH25 pKa = 5.89QIGLLRR31 pKa = 11.84VSGRR35 pKa = 11.84VRR37 pKa = 11.84QRR39 pKa = 11.84IFEE42 pKa = 4.19ILDD45 pKa = 3.15ATEE48 pKa = 5.17RR49 pKa = 11.84DD50 pKa = 3.13IADD53 pKa = 3.44QIKK56 pKa = 10.26RR57 pKa = 11.84RR58 pKa = 11.84LAKK61 pKa = 10.25GATTARR67 pKa = 11.84LEE69 pKa = 4.12ALIKK73 pKa = 10.29AVRR76 pKa = 11.84ALRR79 pKa = 11.84SEE81 pKa = 3.99AWKK84 pKa = 10.6KK85 pKa = 9.79SAEE88 pKa = 4.01VWRR91 pKa = 11.84EE92 pKa = 3.48EE93 pKa = 4.03MLSVARR99 pKa = 11.84EE100 pKa = 3.89EE101 pKa = 4.46PKK103 pKa = 10.69FLSQALRR110 pKa = 11.84TVSPVQLDD118 pKa = 3.83LTLPAADD125 pKa = 4.57LLQSIVTTRR134 pKa = 11.84PFEE137 pKa = 4.33GQTMRR142 pKa = 11.84QWASNIRR149 pKa = 11.84NSDD152 pKa = 3.01LRR154 pKa = 11.84RR155 pKa = 11.84IEE157 pKa = 3.65RR158 pKa = 11.84AIRR161 pKa = 11.84IGVTQGEE168 pKa = 4.52PTNAIARR175 pKa = 11.84RR176 pKa = 11.84VIGTVRR182 pKa = 11.84QRR184 pKa = 11.84GRR186 pKa = 11.84DD187 pKa = 3.16GVTQITRR194 pKa = 11.84RR195 pKa = 11.84NAEE198 pKa = 4.34AITRR202 pKa = 11.84TAINGIANQAKK213 pKa = 10.05RR214 pKa = 11.84EE215 pKa = 4.07FYY217 pKa = 10.23KK218 pKa = 11.21ANATLFEE225 pKa = 4.19EE226 pKa = 4.97EE227 pKa = 4.96IYY229 pKa = 11.05VATLDD234 pKa = 4.0SRR236 pKa = 11.84TTPICRR242 pKa = 11.84SLDD245 pKa = 3.45GQRR248 pKa = 11.84FAIGEE253 pKa = 4.24GPIPPLHH260 pKa = 6.04FNCRR264 pKa = 11.84SLRR267 pKa = 11.84VAAIDD272 pKa = 3.52GDD274 pKa = 4.63AIGSRR279 pKa = 11.84PQRR282 pKa = 11.84QFTQRR287 pKa = 11.84QLLRR291 pKa = 11.84EE292 pKa = 4.15YY293 pKa = 10.92ARR295 pKa = 11.84SRR297 pKa = 11.84GIKK300 pKa = 10.2APTRR304 pKa = 11.84RR305 pKa = 11.84ADD307 pKa = 3.39LPRR310 pKa = 11.84GHH312 pKa = 6.92KK313 pKa = 10.98GEE315 pKa = 4.23FDD317 pKa = 3.17SFARR321 pKa = 11.84TRR323 pKa = 11.84IRR325 pKa = 11.84QLTGTVDD332 pKa = 3.39AKK334 pKa = 11.08VSYY337 pKa = 10.41QDD339 pKa = 2.69WLTRR343 pKa = 11.84QSTEE347 pKa = 3.74FQDD350 pKa = 4.5DD351 pKa = 3.94VLGKK355 pKa = 8.43TRR357 pKa = 11.84ARR359 pKa = 11.84LFRR362 pKa = 11.84KK363 pKa = 10.01GGLRR367 pKa = 11.84LDD369 pKa = 3.51RR370 pKa = 11.84FVNRR374 pKa = 11.84QGDD377 pKa = 4.71EE378 pKa = 3.9IPLADD383 pKa = 4.1LARR386 pKa = 11.84RR387 pKa = 11.84EE388 pKa = 4.11TSAFRR393 pKa = 11.84AAGLDD398 pKa = 3.66PEE400 pKa = 5.61DD401 pKa = 3.89FTT403 pKa = 5.87
MM1 pKa = 7.37PTDD4 pKa = 3.67DD5 pKa = 5.34QIPGMPDD12 pKa = 2.81NVNEE16 pKa = 4.05EE17 pKa = 4.22WLDD20 pKa = 3.31AMIRR24 pKa = 11.84HH25 pKa = 5.89QIGLLRR31 pKa = 11.84VSGRR35 pKa = 11.84VRR37 pKa = 11.84QRR39 pKa = 11.84IFEE42 pKa = 4.19ILDD45 pKa = 3.15ATEE48 pKa = 5.17RR49 pKa = 11.84DD50 pKa = 3.13IADD53 pKa = 3.44QIKK56 pKa = 10.26RR57 pKa = 11.84RR58 pKa = 11.84LAKK61 pKa = 10.25GATTARR67 pKa = 11.84LEE69 pKa = 4.12ALIKK73 pKa = 10.29AVRR76 pKa = 11.84ALRR79 pKa = 11.84SEE81 pKa = 3.99AWKK84 pKa = 10.6KK85 pKa = 9.79SAEE88 pKa = 4.01VWRR91 pKa = 11.84EE92 pKa = 3.48EE93 pKa = 4.03MLSVARR99 pKa = 11.84EE100 pKa = 3.89EE101 pKa = 4.46PKK103 pKa = 10.69FLSQALRR110 pKa = 11.84TVSPVQLDD118 pKa = 3.83LTLPAADD125 pKa = 4.57LLQSIVTTRR134 pKa = 11.84PFEE137 pKa = 4.33GQTMRR142 pKa = 11.84QWASNIRR149 pKa = 11.84NSDD152 pKa = 3.01LRR154 pKa = 11.84RR155 pKa = 11.84IEE157 pKa = 3.65RR158 pKa = 11.84AIRR161 pKa = 11.84IGVTQGEE168 pKa = 4.52PTNAIARR175 pKa = 11.84RR176 pKa = 11.84VIGTVRR182 pKa = 11.84QRR184 pKa = 11.84GRR186 pKa = 11.84DD187 pKa = 3.16GVTQITRR194 pKa = 11.84RR195 pKa = 11.84NAEE198 pKa = 4.34AITRR202 pKa = 11.84TAINGIANQAKK213 pKa = 10.05RR214 pKa = 11.84EE215 pKa = 4.07FYY217 pKa = 10.23KK218 pKa = 11.21ANATLFEE225 pKa = 4.19EE226 pKa = 4.97EE227 pKa = 4.96IYY229 pKa = 11.05VATLDD234 pKa = 4.0SRR236 pKa = 11.84TTPICRR242 pKa = 11.84SLDD245 pKa = 3.45GQRR248 pKa = 11.84FAIGEE253 pKa = 4.24GPIPPLHH260 pKa = 6.04FNCRR264 pKa = 11.84SLRR267 pKa = 11.84VAAIDD272 pKa = 3.52GDD274 pKa = 4.63AIGSRR279 pKa = 11.84PQRR282 pKa = 11.84QFTQRR287 pKa = 11.84QLLRR291 pKa = 11.84EE292 pKa = 4.15YY293 pKa = 10.92ARR295 pKa = 11.84SRR297 pKa = 11.84GIKK300 pKa = 10.2APTRR304 pKa = 11.84RR305 pKa = 11.84ADD307 pKa = 3.39LPRR310 pKa = 11.84GHH312 pKa = 6.92KK313 pKa = 10.98GEE315 pKa = 4.23FDD317 pKa = 3.17SFARR321 pKa = 11.84TRR323 pKa = 11.84IRR325 pKa = 11.84QLTGTVDD332 pKa = 3.39AKK334 pKa = 11.08VSYY337 pKa = 10.41QDD339 pKa = 2.69WLTRR343 pKa = 11.84QSTEE347 pKa = 3.74FQDD350 pKa = 4.5DD351 pKa = 3.94VLGKK355 pKa = 8.43TRR357 pKa = 11.84ARR359 pKa = 11.84LFRR362 pKa = 11.84KK363 pKa = 10.01GGLRR367 pKa = 11.84LDD369 pKa = 3.51RR370 pKa = 11.84FVNRR374 pKa = 11.84QGDD377 pKa = 4.71EE378 pKa = 3.9IPLADD383 pKa = 4.1LARR386 pKa = 11.84RR387 pKa = 11.84EE388 pKa = 4.11TSAFRR393 pKa = 11.84AAGLDD398 pKa = 3.66PEE400 pKa = 5.61DD401 pKa = 3.89FTT403 pKa = 5.87
Molecular weight: 45.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
20205 |
29 |
1152 |
224.5 |
24.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.112 ± 0.361 | 0.97 ± 0.112 |
6.686 ± 0.224 | 6.36 ± 0.271 |
4.078 ± 0.19 | 8.523 ± 0.33 |
1.881 ± 0.204 | 4.85 ± 0.182 |
4.172 ± 0.348 | 7.869 ± 0.207 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.405 ± 0.199 | 4.068 ± 0.18 |
4.147 ± 0.173 | 3.89 ± 0.186 |
6.3 ± 0.304 | 5.914 ± 0.259 |
7.157 ± 0.468 | 7.176 ± 0.164 |
1.891 ± 0.155 | 2.549 ± 0.171 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
