
Faeces associated gemycircularvirus 18
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus equas1; Equine associated gemycircularvirus 1
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
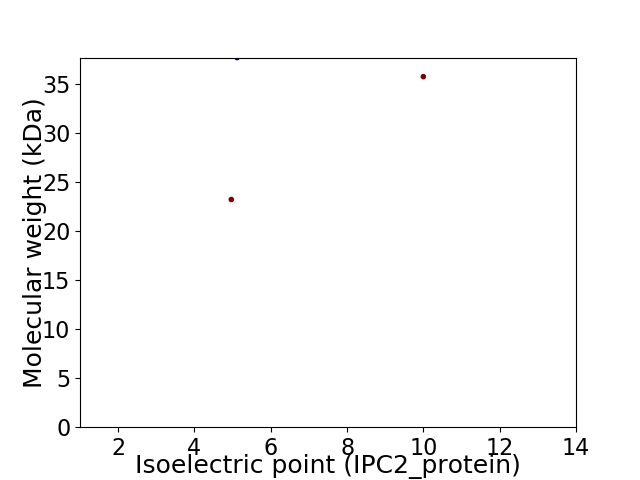
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A160HX62|A0A160HX62_9VIRU Capsid protein OS=Faeces associated gemycircularvirus 18 OX=1843738 PE=4 SV=1
MM1 pKa = 6.76PTFYY5 pKa = 10.64FSARR9 pKa = 11.84YY10 pKa = 9.46VLLTYY15 pKa = 7.98SQCGTLDD22 pKa = 2.99EE23 pKa = 4.76WDD25 pKa = 3.96VLDD28 pKa = 5.56HH29 pKa = 6.74ISSLGAEE36 pKa = 4.45CIIGRR41 pKa = 11.84EE42 pKa = 4.05DD43 pKa = 3.35HH44 pKa = 7.18ADD46 pKa = 3.64GGTHH50 pKa = 6.1LHH52 pKa = 5.74VFVDD56 pKa = 5.19FGHH59 pKa = 6.72KK60 pKa = 9.51KK61 pKa = 8.11QSRR64 pKa = 11.84RR65 pKa = 11.84SGFFDD70 pKa = 3.79VGGKK74 pKa = 9.26HH75 pKa = 6.09PNVVPSRR82 pKa = 11.84GRR84 pKa = 11.84AGEE87 pKa = 4.05GWDD90 pKa = 3.6YY91 pKa = 11.53AVKK94 pKa = 10.58DD95 pKa = 4.04GNVVAGGLPRR105 pKa = 11.84PGSGGLPSVEE115 pKa = 4.91NIWSSIVDD123 pKa = 3.41AQSRR127 pKa = 11.84EE128 pKa = 4.01EE129 pKa = 3.84FLEE132 pKa = 4.05LVRR135 pKa = 11.84ALAPKK140 pKa = 10.32EE141 pKa = 3.61FILRR145 pKa = 11.84HH146 pKa = 5.74KK147 pKa = 10.33EE148 pKa = 3.59LLEE151 pKa = 4.09YY152 pKa = 10.57ADD154 pKa = 3.99RR155 pKa = 11.84YY156 pKa = 7.71YY157 pKa = 11.32AEE159 pKa = 5.19RR160 pKa = 11.84IEE162 pKa = 4.88PYY164 pKa = 10.15VGPDD168 pKa = 3.47GIEE171 pKa = 4.18FEE173 pKa = 4.64LGMVPEE179 pKa = 4.18LAGWGRR185 pKa = 11.84EE186 pKa = 3.95LVEE189 pKa = 4.59ADD191 pKa = 4.15SVTGEE196 pKa = 4.02CPLRR200 pKa = 11.84SGAEE204 pKa = 4.05YY205 pKa = 10.27ISARR209 pKa = 11.84VV210 pKa = 3.16
MM1 pKa = 6.76PTFYY5 pKa = 10.64FSARR9 pKa = 11.84YY10 pKa = 9.46VLLTYY15 pKa = 7.98SQCGTLDD22 pKa = 2.99EE23 pKa = 4.76WDD25 pKa = 3.96VLDD28 pKa = 5.56HH29 pKa = 6.74ISSLGAEE36 pKa = 4.45CIIGRR41 pKa = 11.84EE42 pKa = 4.05DD43 pKa = 3.35HH44 pKa = 7.18ADD46 pKa = 3.64GGTHH50 pKa = 6.1LHH52 pKa = 5.74VFVDD56 pKa = 5.19FGHH59 pKa = 6.72KK60 pKa = 9.51KK61 pKa = 8.11QSRR64 pKa = 11.84RR65 pKa = 11.84SGFFDD70 pKa = 3.79VGGKK74 pKa = 9.26HH75 pKa = 6.09PNVVPSRR82 pKa = 11.84GRR84 pKa = 11.84AGEE87 pKa = 4.05GWDD90 pKa = 3.6YY91 pKa = 11.53AVKK94 pKa = 10.58DD95 pKa = 4.04GNVVAGGLPRR105 pKa = 11.84PGSGGLPSVEE115 pKa = 4.91NIWSSIVDD123 pKa = 3.41AQSRR127 pKa = 11.84EE128 pKa = 4.01EE129 pKa = 3.84FLEE132 pKa = 4.05LVRR135 pKa = 11.84ALAPKK140 pKa = 10.32EE141 pKa = 3.61FILRR145 pKa = 11.84HH146 pKa = 5.74KK147 pKa = 10.33EE148 pKa = 3.59LLEE151 pKa = 4.09YY152 pKa = 10.57ADD154 pKa = 3.99RR155 pKa = 11.84YY156 pKa = 7.71YY157 pKa = 11.32AEE159 pKa = 5.19RR160 pKa = 11.84IEE162 pKa = 4.88PYY164 pKa = 10.15VGPDD168 pKa = 3.47GIEE171 pKa = 4.18FEE173 pKa = 4.64LGMVPEE179 pKa = 4.18LAGWGRR185 pKa = 11.84EE186 pKa = 3.95LVEE189 pKa = 4.59ADD191 pKa = 4.15SVTGEE196 pKa = 4.02CPLRR200 pKa = 11.84SGAEE204 pKa = 4.05YY205 pKa = 10.27ISARR209 pKa = 11.84VV210 pKa = 3.16
Molecular weight: 23.22 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A160HX62|A0A160HX62_9VIRU Capsid protein OS=Faeces associated gemycircularvirus 18 OX=1843738 PE=4 SV=1
MM1 pKa = 7.75AYY3 pKa = 10.17ARR5 pKa = 11.84KK6 pKa = 9.45RR7 pKa = 11.84RR8 pKa = 11.84YY9 pKa = 9.7ARR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 9.23SGSSKK18 pKa = 10.47GKK20 pKa = 8.41RR21 pKa = 11.84TTRR24 pKa = 11.84PSARR28 pKa = 11.84RR29 pKa = 11.84YY30 pKa = 9.61RR31 pKa = 11.84STGRR35 pKa = 11.84TRR37 pKa = 11.84RR38 pKa = 11.84STRR41 pKa = 11.84TRR43 pKa = 11.84PMSKK47 pKa = 10.29KK48 pKa = 10.49SILNTTSRR56 pKa = 11.84KK57 pKa = 9.22KK58 pKa = 10.51RR59 pKa = 11.84NGMLTFSNSTGTGVLASIGQNPLTIAGSAAGNNLGYY95 pKa = 10.7VHH97 pKa = 6.95FRR99 pKa = 11.84PTAMDD104 pKa = 4.39LNDD107 pKa = 4.03NNAAPNSITTMSARR121 pKa = 11.84TSTTCFLRR129 pKa = 11.84GLAEE133 pKa = 4.42NIRR136 pKa = 11.84IEE138 pKa = 4.32TSSGNPWFHH147 pKa = 6.87RR148 pKa = 11.84RR149 pKa = 11.84ICFTSRR155 pKa = 11.84DD156 pKa = 3.77TLFLSVASADD166 pKa = 3.81TPGTDD171 pKa = 2.82RR172 pKa = 11.84AAAASGAIEE181 pKa = 4.6TSNGWQRR188 pKa = 11.84LAANMALDD196 pKa = 3.86TMTGTINNFNGLIFKK211 pKa = 7.6GTQGRR216 pKa = 11.84RR217 pKa = 11.84LGMTSLLHH225 pKa = 6.92LSILHH230 pKa = 6.77ALTSSMTRR238 pKa = 11.84PRR240 pKa = 11.84FTNPGNEE247 pKa = 3.49RR248 pKa = 11.84GVLRR252 pKa = 11.84EE253 pKa = 3.92TKK255 pKa = 9.32IWHH258 pKa = 6.52PMNKK262 pKa = 9.98NIVYY266 pKa = 10.5DD267 pKa = 3.93DD268 pKa = 4.28DD269 pKa = 3.92EE270 pKa = 7.1RR271 pKa = 11.84GDD273 pKa = 4.06KK274 pKa = 10.84YY275 pKa = 10.93ATNNYY280 pKa = 9.98SVTDD284 pKa = 3.5KK285 pKa = 11.19RR286 pKa = 11.84GMGDD290 pKa = 2.99YY291 pKa = 10.64HH292 pKa = 8.78IIDD295 pKa = 4.92LFSQGSSGSTSDD307 pKa = 3.77LLKK310 pKa = 10.32IRR312 pKa = 11.84YY313 pKa = 6.3TSTMYY318 pKa = 9.55WHH320 pKa = 7.09EE321 pKa = 4.17KK322 pKa = 9.5
MM1 pKa = 7.75AYY3 pKa = 10.17ARR5 pKa = 11.84KK6 pKa = 9.45RR7 pKa = 11.84RR8 pKa = 11.84YY9 pKa = 9.7ARR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 9.23SGSSKK18 pKa = 10.47GKK20 pKa = 8.41RR21 pKa = 11.84TTRR24 pKa = 11.84PSARR28 pKa = 11.84RR29 pKa = 11.84YY30 pKa = 9.61RR31 pKa = 11.84STGRR35 pKa = 11.84TRR37 pKa = 11.84RR38 pKa = 11.84STRR41 pKa = 11.84TRR43 pKa = 11.84PMSKK47 pKa = 10.29KK48 pKa = 10.49SILNTTSRR56 pKa = 11.84KK57 pKa = 9.22KK58 pKa = 10.51RR59 pKa = 11.84NGMLTFSNSTGTGVLASIGQNPLTIAGSAAGNNLGYY95 pKa = 10.7VHH97 pKa = 6.95FRR99 pKa = 11.84PTAMDD104 pKa = 4.39LNDD107 pKa = 4.03NNAAPNSITTMSARR121 pKa = 11.84TSTTCFLRR129 pKa = 11.84GLAEE133 pKa = 4.42NIRR136 pKa = 11.84IEE138 pKa = 4.32TSSGNPWFHH147 pKa = 6.87RR148 pKa = 11.84RR149 pKa = 11.84ICFTSRR155 pKa = 11.84DD156 pKa = 3.77TLFLSVASADD166 pKa = 3.81TPGTDD171 pKa = 2.82RR172 pKa = 11.84AAAASGAIEE181 pKa = 4.6TSNGWQRR188 pKa = 11.84LAANMALDD196 pKa = 3.86TMTGTINNFNGLIFKK211 pKa = 7.6GTQGRR216 pKa = 11.84RR217 pKa = 11.84LGMTSLLHH225 pKa = 6.92LSILHH230 pKa = 6.77ALTSSMTRR238 pKa = 11.84PRR240 pKa = 11.84FTNPGNEE247 pKa = 3.49RR248 pKa = 11.84GVLRR252 pKa = 11.84EE253 pKa = 3.92TKK255 pKa = 9.32IWHH258 pKa = 6.52PMNKK262 pKa = 9.98NIVYY266 pKa = 10.5DD267 pKa = 3.93DD268 pKa = 4.28DD269 pKa = 3.92EE270 pKa = 7.1RR271 pKa = 11.84GDD273 pKa = 4.06KK274 pKa = 10.84YY275 pKa = 10.93ATNNYY280 pKa = 9.98SVTDD284 pKa = 3.5KK285 pKa = 11.19RR286 pKa = 11.84GMGDD290 pKa = 2.99YY291 pKa = 10.64HH292 pKa = 8.78IIDD295 pKa = 4.92LFSQGSSGSTSDD307 pKa = 3.77LLKK310 pKa = 10.32IRR312 pKa = 11.84YY313 pKa = 6.3TSTMYY318 pKa = 9.55WHH320 pKa = 7.09EE321 pKa = 4.17KK322 pKa = 9.5
Molecular weight: 35.75 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
870 |
210 |
338 |
290.0 |
32.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.356 ± 0.186 | 1.034 ± 0.197 |
5.862 ± 0.719 | 5.862 ± 1.772 |
3.793 ± 0.315 | 9.885 ± 0.806 |
2.529 ± 0.276 | 4.713 ± 0.156 |
3.908 ± 0.412 | 8.391 ± 0.603 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.299 ± 0.678 | 3.678 ± 1.579 |
4.483 ± 0.627 | 1.609 ± 0.223 |
8.391 ± 1.13 | 8.276 ± 1.042 |
6.207 ± 2.555 | 6.092 ± 1.924 |
1.954 ± 0.367 | 3.678 ± 0.28 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
