
Caligus rogercresseyi rhabdovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Caligrhavirus; Caligus caligrhavirus
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
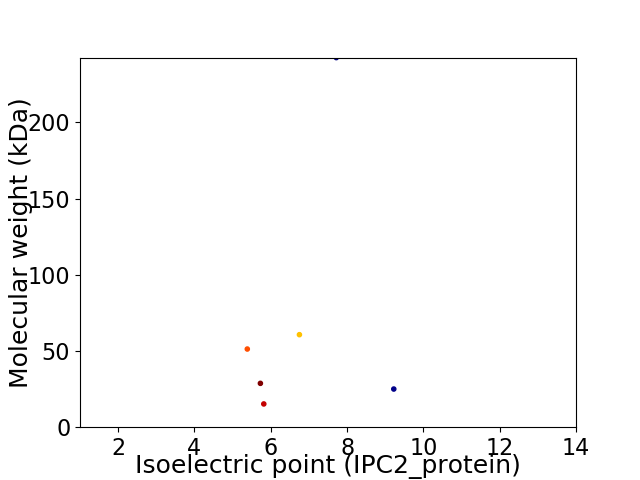
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L2YZV1|A0A1L2YZV1_9RHAB Glycoprotein OS=Caligus rogercresseyi rhabdovirus OX=1921414 GN=G PE=4 SV=1
MM1 pKa = 8.22DD2 pKa = 6.52DD3 pKa = 5.64DD4 pKa = 5.35IIHH7 pKa = 6.85FKK9 pKa = 8.87TGKK12 pKa = 9.68VFNLLAPVEE21 pKa = 4.29NLPTQYY27 pKa = 10.41PATWFQQEE35 pKa = 4.12KK36 pKa = 9.66KK37 pKa = 10.39KK38 pKa = 10.66PYY40 pKa = 8.78LTLRR44 pKa = 11.84LPQSLIAEE52 pKa = 4.74PARR55 pKa = 11.84IVALLEE61 pKa = 3.84AQMQIGRR68 pKa = 11.84VTPDD72 pKa = 2.98VLAFGYY78 pKa = 10.51KK79 pKa = 9.99FLLDD83 pKa = 3.81RR84 pKa = 11.84EE85 pKa = 4.47VLTRR89 pKa = 11.84DD90 pKa = 3.03GPAWVSFDD98 pKa = 3.75QQITHH103 pKa = 6.01EE104 pKa = 4.43NKK106 pKa = 10.13DD107 pKa = 3.73RR108 pKa = 11.84LDD110 pKa = 4.93GILTMTMQAEE120 pKa = 4.81DD121 pKa = 3.95GLEE124 pKa = 4.01MKK126 pKa = 10.01EE127 pKa = 4.22GKK129 pKa = 7.51PTLVDD134 pKa = 3.21TLYY137 pKa = 11.47VNLVVCGAIVRR148 pKa = 11.84RR149 pKa = 11.84DD150 pKa = 3.29RR151 pKa = 11.84VANVAGKK158 pKa = 10.14DD159 pKa = 3.38RR160 pKa = 11.84ITQIVTRR167 pKa = 11.84AVQSSMGIDD176 pKa = 2.88ISGYY180 pKa = 9.22LGSVQVTTNATTTKK194 pKa = 10.21IMAAVDD200 pKa = 3.25MYY202 pKa = 11.14LYY204 pKa = 10.6NRR206 pKa = 11.84RR207 pKa = 11.84DD208 pKa = 3.08SMYY211 pKa = 10.62GFLRR215 pKa = 11.84LGTVCTRR222 pKa = 11.84YY223 pKa = 9.8RR224 pKa = 11.84DD225 pKa = 3.85CTVLSEE231 pKa = 4.39FVLLEE236 pKa = 4.52DD237 pKa = 5.58LSQTKK242 pKa = 10.26CKK244 pKa = 10.12EE245 pKa = 4.12AISWILSPIVSDD257 pKa = 3.69EE258 pKa = 5.0LISTMEE264 pKa = 4.68PGQEE268 pKa = 3.91THH270 pKa = 6.63SVGSYY275 pKa = 8.44MPYY278 pKa = 10.38MMDD281 pKa = 3.35LQLSTRR287 pKa = 11.84SPYY290 pKa = 9.27STSEE294 pKa = 3.64NPKK297 pKa = 7.58YY298 pKa = 9.29HH299 pKa = 5.15TWIHH303 pKa = 5.32IVGTLLHH310 pKa = 7.0KK311 pKa = 10.48DD312 pKa = 3.31RR313 pKa = 11.84CRR315 pKa = 11.84KK316 pKa = 9.42AAVIEE321 pKa = 3.93EE322 pKa = 4.12ALIGRR327 pKa = 11.84LFKK330 pKa = 10.85GAALAAYY337 pKa = 8.89RR338 pKa = 11.84WTEE341 pKa = 3.56SDD343 pKa = 3.19EE344 pKa = 4.16VVQVFGRR351 pKa = 11.84MRR353 pKa = 11.84DD354 pKa = 3.52KK355 pKa = 11.12VIEE358 pKa = 4.04EE359 pKa = 3.96SAAEE363 pKa = 3.83IRR365 pKa = 11.84RR366 pKa = 11.84SQLEE370 pKa = 3.95ANFLDD375 pKa = 4.38GTDD378 pKa = 4.23PEE380 pKa = 4.42EE381 pKa = 4.86DD382 pKa = 3.64SPGAYY387 pKa = 10.01ADD389 pKa = 3.7WARR392 pKa = 11.84DD393 pKa = 3.45EE394 pKa = 4.31YY395 pKa = 10.52PRR397 pKa = 11.84KK398 pKa = 10.04LFQFIDD404 pKa = 3.46NTLNGQFMVIRR415 pKa = 11.84KK416 pKa = 9.66GSMGDD421 pKa = 3.06FVRR424 pKa = 11.84NHH426 pKa = 5.31WAGHH430 pKa = 6.01RR431 pKa = 11.84DD432 pKa = 3.3SLVPEE437 pKa = 4.49KK438 pKa = 11.05FEE440 pKa = 4.14LPPWNGSIFPNAA452 pKa = 3.87
MM1 pKa = 8.22DD2 pKa = 6.52DD3 pKa = 5.64DD4 pKa = 5.35IIHH7 pKa = 6.85FKK9 pKa = 8.87TGKK12 pKa = 9.68VFNLLAPVEE21 pKa = 4.29NLPTQYY27 pKa = 10.41PATWFQQEE35 pKa = 4.12KK36 pKa = 9.66KK37 pKa = 10.39KK38 pKa = 10.66PYY40 pKa = 8.78LTLRR44 pKa = 11.84LPQSLIAEE52 pKa = 4.74PARR55 pKa = 11.84IVALLEE61 pKa = 3.84AQMQIGRR68 pKa = 11.84VTPDD72 pKa = 2.98VLAFGYY78 pKa = 10.51KK79 pKa = 9.99FLLDD83 pKa = 3.81RR84 pKa = 11.84EE85 pKa = 4.47VLTRR89 pKa = 11.84DD90 pKa = 3.03GPAWVSFDD98 pKa = 3.75QQITHH103 pKa = 6.01EE104 pKa = 4.43NKK106 pKa = 10.13DD107 pKa = 3.73RR108 pKa = 11.84LDD110 pKa = 4.93GILTMTMQAEE120 pKa = 4.81DD121 pKa = 3.95GLEE124 pKa = 4.01MKK126 pKa = 10.01EE127 pKa = 4.22GKK129 pKa = 7.51PTLVDD134 pKa = 3.21TLYY137 pKa = 11.47VNLVVCGAIVRR148 pKa = 11.84RR149 pKa = 11.84DD150 pKa = 3.29RR151 pKa = 11.84VANVAGKK158 pKa = 10.14DD159 pKa = 3.38RR160 pKa = 11.84ITQIVTRR167 pKa = 11.84AVQSSMGIDD176 pKa = 2.88ISGYY180 pKa = 9.22LGSVQVTTNATTTKK194 pKa = 10.21IMAAVDD200 pKa = 3.25MYY202 pKa = 11.14LYY204 pKa = 10.6NRR206 pKa = 11.84RR207 pKa = 11.84DD208 pKa = 3.08SMYY211 pKa = 10.62GFLRR215 pKa = 11.84LGTVCTRR222 pKa = 11.84YY223 pKa = 9.8RR224 pKa = 11.84DD225 pKa = 3.85CTVLSEE231 pKa = 4.39FVLLEE236 pKa = 4.52DD237 pKa = 5.58LSQTKK242 pKa = 10.26CKK244 pKa = 10.12EE245 pKa = 4.12AISWILSPIVSDD257 pKa = 3.69EE258 pKa = 5.0LISTMEE264 pKa = 4.68PGQEE268 pKa = 3.91THH270 pKa = 6.63SVGSYY275 pKa = 8.44MPYY278 pKa = 10.38MMDD281 pKa = 3.35LQLSTRR287 pKa = 11.84SPYY290 pKa = 9.27STSEE294 pKa = 3.64NPKK297 pKa = 7.58YY298 pKa = 9.29HH299 pKa = 5.15TWIHH303 pKa = 5.32IVGTLLHH310 pKa = 7.0KK311 pKa = 10.48DD312 pKa = 3.31RR313 pKa = 11.84CRR315 pKa = 11.84KK316 pKa = 9.42AAVIEE321 pKa = 3.93EE322 pKa = 4.12ALIGRR327 pKa = 11.84LFKK330 pKa = 10.85GAALAAYY337 pKa = 8.89RR338 pKa = 11.84WTEE341 pKa = 3.56SDD343 pKa = 3.19EE344 pKa = 4.16VVQVFGRR351 pKa = 11.84MRR353 pKa = 11.84DD354 pKa = 3.52KK355 pKa = 11.12VIEE358 pKa = 4.04EE359 pKa = 3.96SAAEE363 pKa = 3.83IRR365 pKa = 11.84RR366 pKa = 11.84SQLEE370 pKa = 3.95ANFLDD375 pKa = 4.38GTDD378 pKa = 4.23PEE380 pKa = 4.42EE381 pKa = 4.86DD382 pKa = 3.64SPGAYY387 pKa = 10.01ADD389 pKa = 3.7WARR392 pKa = 11.84DD393 pKa = 3.45EE394 pKa = 4.31YY395 pKa = 10.52PRR397 pKa = 11.84KK398 pKa = 10.04LFQFIDD404 pKa = 3.46NTLNGQFMVIRR415 pKa = 11.84KK416 pKa = 9.66GSMGDD421 pKa = 3.06FVRR424 pKa = 11.84NHH426 pKa = 5.31WAGHH430 pKa = 6.01RR431 pKa = 11.84DD432 pKa = 3.3SLVPEE437 pKa = 4.49KK438 pKa = 11.05FEE440 pKa = 4.14LPPWNGSIFPNAA452 pKa = 3.87
Molecular weight: 51.27 kDa
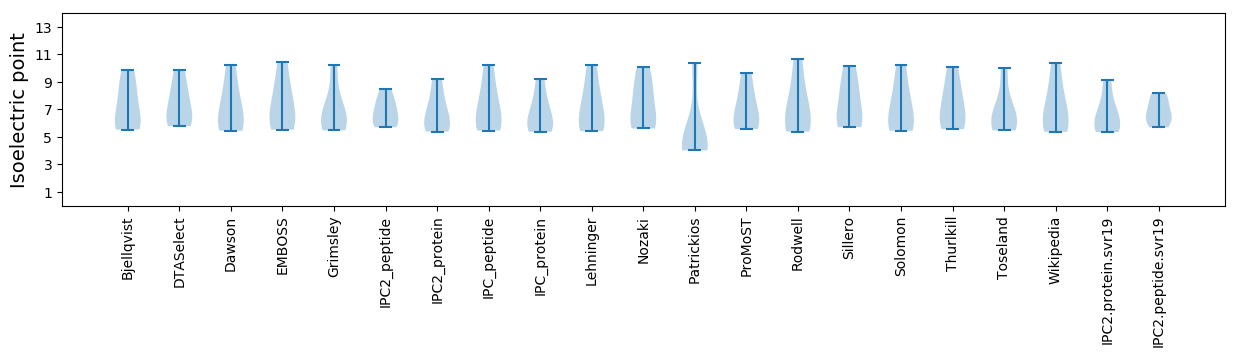
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L2YZU8|A0A1L2YZU8_9RHAB Uncharacterized protein OS=Caligus rogercresseyi rhabdovirus OX=1921414 PE=4 SV=1
MM1 pKa = 7.39KK2 pKa = 10.35KK3 pKa = 9.71IFRR6 pKa = 11.84SKK8 pKa = 10.71RR9 pKa = 11.84LGSTPVASAPTVPGPWRR26 pKa = 11.84ISPQEE31 pKa = 4.1DD32 pKa = 3.27LSKK35 pKa = 10.75RR36 pKa = 11.84GASAGPSKK44 pKa = 9.99TKK46 pKa = 10.66GQGSPQCNCRR56 pKa = 11.84EE57 pKa = 3.77VKK59 pKa = 10.55LRR61 pKa = 11.84IKK63 pKa = 10.26GHH65 pKa = 5.46LSIRR69 pKa = 11.84LGSSPRR75 pKa = 11.84TRR77 pKa = 11.84EE78 pKa = 3.61QAQYY82 pKa = 11.48ACTNFLDD89 pKa = 3.85QYY91 pKa = 10.88GEE93 pKa = 4.36SIEE96 pKa = 4.67KK97 pKa = 10.6SGLYY101 pKa = 5.68WTAVIILARR110 pKa = 11.84KK111 pKa = 7.76LQSVHH116 pKa = 6.73PGGGNSDD123 pKa = 3.67YY124 pKa = 10.64TASLDD129 pKa = 3.79EE130 pKa = 5.22NISILTTIASDD141 pKa = 4.19LLNDD145 pKa = 3.52IPKK148 pKa = 10.66LEE150 pKa = 4.12MSYY153 pKa = 8.98GTQEE157 pKa = 4.51GISGRR162 pKa = 11.84FGIISIQLTPTRR174 pKa = 11.84SPALPLRR181 pKa = 11.84AVLAKK186 pKa = 10.26IKK188 pKa = 10.73LPAEE192 pKa = 4.28KK193 pKa = 9.37FIKK196 pKa = 9.54MAEE199 pKa = 4.38TFGVTYY205 pKa = 10.26EE206 pKa = 4.67LIQGKK211 pKa = 9.17IVLHH215 pKa = 5.58TTTQSRR221 pKa = 11.84SHH223 pKa = 6.29LRR225 pKa = 11.84EE226 pKa = 3.76VVLL229 pKa = 4.9
MM1 pKa = 7.39KK2 pKa = 10.35KK3 pKa = 9.71IFRR6 pKa = 11.84SKK8 pKa = 10.71RR9 pKa = 11.84LGSTPVASAPTVPGPWRR26 pKa = 11.84ISPQEE31 pKa = 4.1DD32 pKa = 3.27LSKK35 pKa = 10.75RR36 pKa = 11.84GASAGPSKK44 pKa = 9.99TKK46 pKa = 10.66GQGSPQCNCRR56 pKa = 11.84EE57 pKa = 3.77VKK59 pKa = 10.55LRR61 pKa = 11.84IKK63 pKa = 10.26GHH65 pKa = 5.46LSIRR69 pKa = 11.84LGSSPRR75 pKa = 11.84TRR77 pKa = 11.84EE78 pKa = 3.61QAQYY82 pKa = 11.48ACTNFLDD89 pKa = 3.85QYY91 pKa = 10.88GEE93 pKa = 4.36SIEE96 pKa = 4.67KK97 pKa = 10.6SGLYY101 pKa = 5.68WTAVIILARR110 pKa = 11.84KK111 pKa = 7.76LQSVHH116 pKa = 6.73PGGGNSDD123 pKa = 3.67YY124 pKa = 10.64TASLDD129 pKa = 3.79EE130 pKa = 5.22NISILTTIASDD141 pKa = 4.19LLNDD145 pKa = 3.52IPKK148 pKa = 10.66LEE150 pKa = 4.12MSYY153 pKa = 8.98GTQEE157 pKa = 4.51GISGRR162 pKa = 11.84FGIISIQLTPTRR174 pKa = 11.84SPALPLRR181 pKa = 11.84AVLAKK186 pKa = 10.26IKK188 pKa = 10.73LPAEE192 pKa = 4.28KK193 pKa = 9.37FIKK196 pKa = 9.54MAEE199 pKa = 4.38TFGVTYY205 pKa = 10.26EE206 pKa = 4.67LIQGKK211 pKa = 9.17IVLHH215 pKa = 5.58TTTQSRR221 pKa = 11.84SHH223 pKa = 6.29LRR225 pKa = 11.84EE226 pKa = 3.76VVLL229 pKa = 4.9
Molecular weight: 25.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3737 |
136 |
2130 |
622.8 |
70.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.272 ± 0.465 | 1.525 ± 0.512 |
5.727 ± 0.313 | 6.342 ± 0.345 |
3.746 ± 0.395 | 6.396 ± 0.31 |
2.275 ± 0.272 | 5.941 ± 0.434 |
5.539 ± 0.67 | 10.891 ± 0.758 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.034 ± 0.323 | 3.72 ± 0.286 |
4.977 ± 0.376 | 3.372 ± 0.361 |
6.904 ± 0.498 | 8.188 ± 0.46 |
6.529 ± 0.595 | 5.593 ± 0.316 |
1.82 ± 0.237 | 3.211 ± 0.256 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
