
bacterium SCGC AG-212-C10
Taxonomy: cellular organisms; Bacteria; unclassified Bacteria
Average proteome isoelectric point is 5.92
Get precalculated fractions of proteins
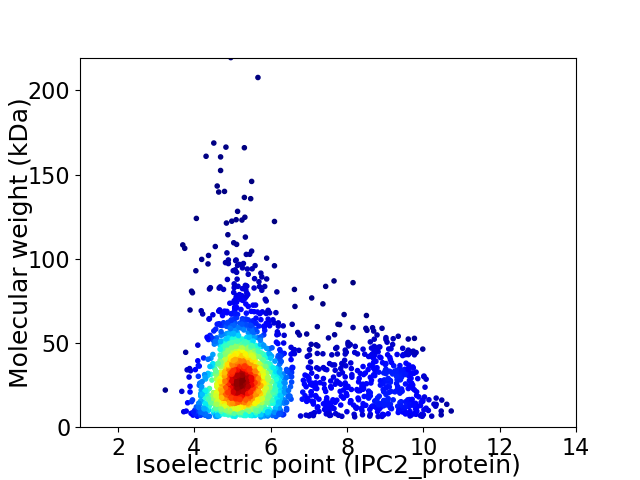
Virtual 2D-PAGE plot for 2196 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A177PYL2|A0A177PYL2_9BACT HTH merR-type domain-containing protein OS=bacterium SCGC AG-212-C10 OX=1799370 GN=AYO38_07020 PE=4 SV=1
MM1 pKa = 7.71LDD3 pKa = 3.35VFGTAVAISGNTIVVSGYY21 pKa = 11.16GEE23 pKa = 4.49DD24 pKa = 3.73SSTTGVNGDD33 pKa = 3.63QEE35 pKa = 5.13DD36 pKa = 4.05NSSATSFAAYY46 pKa = 10.05VFVRR50 pKa = 11.84NGATWVQQAYY60 pKa = 10.55LKK62 pKa = 10.87ASNNDD67 pKa = 3.12AGDD70 pKa = 3.56FFGYY74 pKa = 9.87SVAVSVDD81 pKa = 3.79TIVVGAINEE90 pKa = 4.07ASKK93 pKa = 9.99ATGVNHH99 pKa = 7.01EE100 pKa = 5.09EE101 pKa = 4.68GDD103 pKa = 3.83DD104 pKa = 3.81SAPEE108 pKa = 3.7AGAAYY113 pKa = 10.45VFVRR117 pKa = 11.84DD118 pKa = 3.7GTTWTQQAYY127 pKa = 10.31LKK129 pKa = 10.82ASNTDD134 pKa = 2.83AGDD137 pKa = 3.4RR138 pKa = 11.84FSYY141 pKa = 10.93VSVSGDD147 pKa = 3.59TLVVGADD154 pKa = 4.09LEE156 pKa = 4.42ASHH159 pKa = 7.22ASGKK163 pKa = 10.2NGNQADD169 pKa = 3.84NSAPQAGAAYY179 pKa = 10.24VFVRR183 pKa = 11.84SNQVWTQRR191 pKa = 11.84AYY193 pKa = 11.26LKK195 pKa = 10.31ATVAEE200 pKa = 4.33ANSFFGFSVAISGNAVLVGAYY221 pKa = 9.66GQDD224 pKa = 3.34VTVGGVGAAYY234 pKa = 9.79VYY236 pKa = 10.5PFEE239 pKa = 4.51QDD241 pKa = 3.33VFACDD246 pKa = 3.65GTLDD250 pKa = 3.95SADD253 pKa = 3.22ALAIFRR259 pKa = 11.84ALAGLTGAPTPTAPCTGDD277 pKa = 3.23YY278 pKa = 11.17DD279 pKa = 3.96EE280 pKa = 6.22SGDD283 pKa = 4.01VNLQDD288 pKa = 3.27VVAVLRR294 pKa = 11.84QLAGMTNLEE303 pKa = 4.25PQLQSLMFAPSPVAAGGSTTLSFTVTNADD332 pKa = 3.1
MM1 pKa = 7.71LDD3 pKa = 3.35VFGTAVAISGNTIVVSGYY21 pKa = 11.16GEE23 pKa = 4.49DD24 pKa = 3.73SSTTGVNGDD33 pKa = 3.63QEE35 pKa = 5.13DD36 pKa = 4.05NSSATSFAAYY46 pKa = 10.05VFVRR50 pKa = 11.84NGATWVQQAYY60 pKa = 10.55LKK62 pKa = 10.87ASNNDD67 pKa = 3.12AGDD70 pKa = 3.56FFGYY74 pKa = 9.87SVAVSVDD81 pKa = 3.79TIVVGAINEE90 pKa = 4.07ASKK93 pKa = 9.99ATGVNHH99 pKa = 7.01EE100 pKa = 5.09EE101 pKa = 4.68GDD103 pKa = 3.83DD104 pKa = 3.81SAPEE108 pKa = 3.7AGAAYY113 pKa = 10.45VFVRR117 pKa = 11.84DD118 pKa = 3.7GTTWTQQAYY127 pKa = 10.31LKK129 pKa = 10.82ASNTDD134 pKa = 2.83AGDD137 pKa = 3.4RR138 pKa = 11.84FSYY141 pKa = 10.93VSVSGDD147 pKa = 3.59TLVVGADD154 pKa = 4.09LEE156 pKa = 4.42ASHH159 pKa = 7.22ASGKK163 pKa = 10.2NGNQADD169 pKa = 3.84NSAPQAGAAYY179 pKa = 10.24VFVRR183 pKa = 11.84SNQVWTQRR191 pKa = 11.84AYY193 pKa = 11.26LKK195 pKa = 10.31ATVAEE200 pKa = 4.33ANSFFGFSVAISGNAVLVGAYY221 pKa = 9.66GQDD224 pKa = 3.34VTVGGVGAAYY234 pKa = 9.79VYY236 pKa = 10.5PFEE239 pKa = 4.51QDD241 pKa = 3.33VFACDD246 pKa = 3.65GTLDD250 pKa = 3.95SADD253 pKa = 3.22ALAIFRR259 pKa = 11.84ALAGLTGAPTPTAPCTGDD277 pKa = 3.23YY278 pKa = 11.17DD279 pKa = 3.96EE280 pKa = 6.22SGDD283 pKa = 4.01VNLQDD288 pKa = 3.27VVAVLRR294 pKa = 11.84QLAGMTNLEE303 pKa = 4.25PQLQSLMFAPSPVAAGGSTTLSFTVTNADD332 pKa = 3.1
Molecular weight: 34.07 kDa
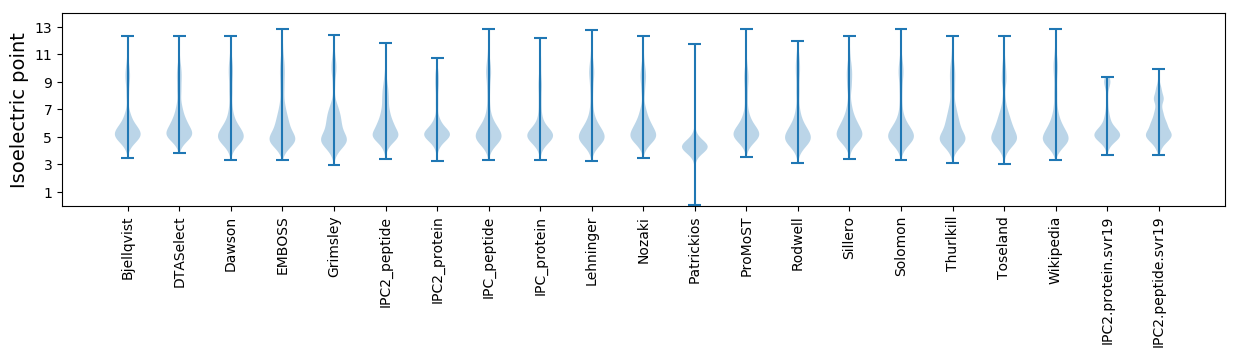
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A177PZ98|A0A177PZ98_9BACT Glyoxalase OS=bacterium SCGC AG-212-C10 OX=1799370 GN=AYO38_05980 PE=4 SV=1
MM1 pKa = 7.14NLSVATRR8 pKa = 11.84IEE10 pKa = 4.31NNEE13 pKa = 4.09SPSLQRR19 pKa = 11.84FEE21 pKa = 4.5IPGQIVAMPAWKK33 pKa = 8.79RR34 pKa = 11.84TMDD37 pKa = 3.12IFGASVGLLMLSPLLVLVSAAVMLDD62 pKa = 3.23SRR64 pKa = 11.84GAPFFRR70 pKa = 11.84QRR72 pKa = 11.84RR73 pKa = 11.84IGRR76 pKa = 11.84GGMPFLCWKK85 pKa = 9.04FRR87 pKa = 11.84SMHH90 pKa = 5.53QGAEE94 pKa = 3.83RR95 pKa = 11.84LQAQLNAYY103 pKa = 9.75NEE105 pKa = 4.25ANGCIFKK112 pKa = 9.99MRR114 pKa = 11.84DD115 pKa = 3.07DD116 pKa = 3.81PRR118 pKa = 11.84RR119 pKa = 11.84TRR121 pKa = 11.84VGKK124 pKa = 8.35VLRR127 pKa = 11.84RR128 pKa = 11.84TSLDD132 pKa = 3.51EE133 pKa = 4.45LPQLINVLLGQMSLVGPRR151 pKa = 11.84PPVIPEE157 pKa = 3.81VVQYY161 pKa = 10.88QPDD164 pKa = 3.7EE165 pKa = 4.12LRR167 pKa = 11.84RR168 pKa = 11.84LAMVPGMTGLWQVSRR183 pKa = 11.84RR184 pKa = 11.84GAFEE188 pKa = 3.75FSEE191 pKa = 4.48MVALDD196 pKa = 3.46VEE198 pKa = 4.52YY199 pKa = 11.16GEE201 pKa = 5.11RR202 pKa = 11.84LTFRR206 pKa = 11.84RR207 pKa = 11.84DD208 pKa = 2.95IQILLRR214 pKa = 11.84TIPTVMFGRR223 pKa = 11.84GSYY226 pKa = 10.22
MM1 pKa = 7.14NLSVATRR8 pKa = 11.84IEE10 pKa = 4.31NNEE13 pKa = 4.09SPSLQRR19 pKa = 11.84FEE21 pKa = 4.5IPGQIVAMPAWKK33 pKa = 8.79RR34 pKa = 11.84TMDD37 pKa = 3.12IFGASVGLLMLSPLLVLVSAAVMLDD62 pKa = 3.23SRR64 pKa = 11.84GAPFFRR70 pKa = 11.84QRR72 pKa = 11.84RR73 pKa = 11.84IGRR76 pKa = 11.84GGMPFLCWKK85 pKa = 9.04FRR87 pKa = 11.84SMHH90 pKa = 5.53QGAEE94 pKa = 3.83RR95 pKa = 11.84LQAQLNAYY103 pKa = 9.75NEE105 pKa = 4.25ANGCIFKK112 pKa = 9.99MRR114 pKa = 11.84DD115 pKa = 3.07DD116 pKa = 3.81PRR118 pKa = 11.84RR119 pKa = 11.84TRR121 pKa = 11.84VGKK124 pKa = 8.35VLRR127 pKa = 11.84RR128 pKa = 11.84TSLDD132 pKa = 3.51EE133 pKa = 4.45LPQLINVLLGQMSLVGPRR151 pKa = 11.84PPVIPEE157 pKa = 3.81VVQYY161 pKa = 10.88QPDD164 pKa = 3.7EE165 pKa = 4.12LRR167 pKa = 11.84RR168 pKa = 11.84LAMVPGMTGLWQVSRR183 pKa = 11.84RR184 pKa = 11.84GAFEE188 pKa = 3.75FSEE191 pKa = 4.48MVALDD196 pKa = 3.46VEE198 pKa = 4.52YY199 pKa = 11.16GEE201 pKa = 5.11RR202 pKa = 11.84LTFRR206 pKa = 11.84RR207 pKa = 11.84DD208 pKa = 2.95IQILLRR214 pKa = 11.84TIPTVMFGRR223 pKa = 11.84GSYY226 pKa = 10.22
Molecular weight: 25.67 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
665205 |
55 |
2017 |
302.9 |
32.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.062 ± 0.079 | 0.854 ± 0.017 |
5.753 ± 0.04 | 5.858 ± 0.06 |
3.612 ± 0.036 | 8.99 ± 0.059 |
2.002 ± 0.026 | 4.933 ± 0.037 |
2.583 ± 0.043 | 9.394 ± 0.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.287 ± 0.027 | 2.816 ± 0.045 |
5.388 ± 0.042 | 3.123 ± 0.028 |
6.886 ± 0.06 | 5.853 ± 0.047 |
5.914 ± 0.059 | 7.885 ± 0.043 |
1.448 ± 0.021 | 2.36 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
