
Watermelon virus A
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Tymovirales; Betaflexiviridae; Trivirinae; Wamavirus
Average proteome isoelectric point is 6.84
Get precalculated fractions of proteins
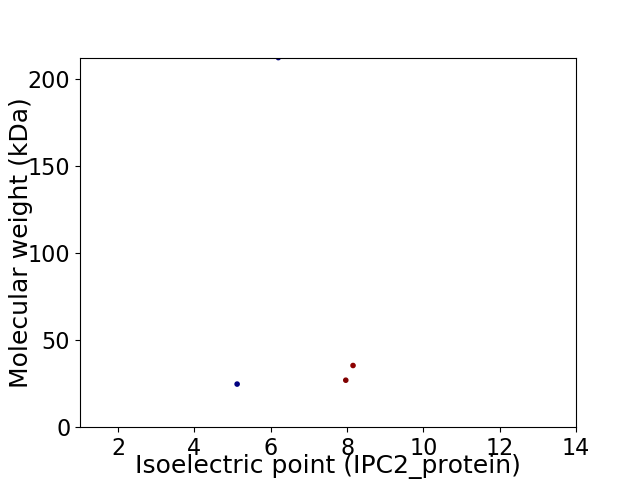
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1W5W6E2|A0A1W5W6E2_9VIRU Replication-associated polyprotein OS=Watermelon virus A OX=1978413 PE=4 SV=1
MM1 pKa = 7.87EE2 pKa = 6.25DD3 pKa = 4.66LPDD6 pKa = 4.0KK7 pKa = 10.84APRR10 pKa = 11.84DD11 pKa = 3.61ILEE14 pKa = 4.09KK15 pKa = 10.22HH16 pKa = 4.68LHH18 pKa = 5.11EE19 pKa = 4.65RR20 pKa = 11.84EE21 pKa = 4.07CFKK24 pKa = 10.9FLEE27 pKa = 4.21QSFDD31 pKa = 3.69AGDD34 pKa = 3.55KK35 pKa = 10.69GQIPSILVKK44 pKa = 9.52LTRR47 pKa = 11.84VNVGFYY53 pKa = 10.68EE54 pKa = 4.01FLEE57 pKa = 4.48EE58 pKa = 4.54VFTEE62 pKa = 4.2SSISYY67 pKa = 9.86GLSGLEE73 pKa = 4.7KK74 pKa = 10.94GFDD77 pKa = 3.55SLKK80 pKa = 11.17AKK82 pKa = 9.72IDD84 pKa = 3.99KK85 pKa = 10.58INSEE89 pKa = 3.99VSEE92 pKa = 4.31MKK94 pKa = 10.46RR95 pKa = 11.84KK96 pKa = 9.85FEE98 pKa = 3.94NLEE101 pKa = 3.78IKK103 pKa = 10.41LVEE106 pKa = 4.31SQSSEE111 pKa = 4.18EE112 pKa = 4.1EE113 pKa = 4.2GSPDD117 pKa = 3.14VYY119 pKa = 9.55EE120 pKa = 4.51TKK122 pKa = 10.54FSDD125 pKa = 3.41MRR127 pKa = 11.84CDD129 pKa = 3.22RR130 pKa = 11.84KK131 pKa = 9.99GTFLYY136 pKa = 10.31TFEE139 pKa = 4.49GQVNARR145 pKa = 11.84ILSDD149 pKa = 3.75FKK151 pKa = 11.11PFEE154 pKa = 4.43CEE156 pKa = 3.52SGKK159 pKa = 10.5FLVHH163 pKa = 6.87KK164 pKa = 10.69YY165 pKa = 8.1VDD167 pKa = 3.6KK168 pKa = 11.45YY169 pKa = 11.83EE170 pKa = 4.3MIFTHH175 pKa = 7.04LGQVMGYY182 pKa = 9.35FEE184 pKa = 5.56IEE186 pKa = 4.17TNKK189 pKa = 10.34EE190 pKa = 3.9LTFGTLIHH198 pKa = 6.65FLRR201 pKa = 11.84EE202 pKa = 3.78KK203 pKa = 11.13YY204 pKa = 10.64SFNKK208 pKa = 10.25LFIII212 pKa = 5.22
MM1 pKa = 7.87EE2 pKa = 6.25DD3 pKa = 4.66LPDD6 pKa = 4.0KK7 pKa = 10.84APRR10 pKa = 11.84DD11 pKa = 3.61ILEE14 pKa = 4.09KK15 pKa = 10.22HH16 pKa = 4.68LHH18 pKa = 5.11EE19 pKa = 4.65RR20 pKa = 11.84EE21 pKa = 4.07CFKK24 pKa = 10.9FLEE27 pKa = 4.21QSFDD31 pKa = 3.69AGDD34 pKa = 3.55KK35 pKa = 10.69GQIPSILVKK44 pKa = 9.52LTRR47 pKa = 11.84VNVGFYY53 pKa = 10.68EE54 pKa = 4.01FLEE57 pKa = 4.48EE58 pKa = 4.54VFTEE62 pKa = 4.2SSISYY67 pKa = 9.86GLSGLEE73 pKa = 4.7KK74 pKa = 10.94GFDD77 pKa = 3.55SLKK80 pKa = 11.17AKK82 pKa = 9.72IDD84 pKa = 3.99KK85 pKa = 10.58INSEE89 pKa = 3.99VSEE92 pKa = 4.31MKK94 pKa = 10.46RR95 pKa = 11.84KK96 pKa = 9.85FEE98 pKa = 3.94NLEE101 pKa = 3.78IKK103 pKa = 10.41LVEE106 pKa = 4.31SQSSEE111 pKa = 4.18EE112 pKa = 4.1EE113 pKa = 4.2GSPDD117 pKa = 3.14VYY119 pKa = 9.55EE120 pKa = 4.51TKK122 pKa = 10.54FSDD125 pKa = 3.41MRR127 pKa = 11.84CDD129 pKa = 3.22RR130 pKa = 11.84KK131 pKa = 9.99GTFLYY136 pKa = 10.31TFEE139 pKa = 4.49GQVNARR145 pKa = 11.84ILSDD149 pKa = 3.75FKK151 pKa = 11.11PFEE154 pKa = 4.43CEE156 pKa = 3.52SGKK159 pKa = 10.5FLVHH163 pKa = 6.87KK164 pKa = 10.69YY165 pKa = 8.1VDD167 pKa = 3.6KK168 pKa = 11.45YY169 pKa = 11.83EE170 pKa = 4.3MIFTHH175 pKa = 7.04LGQVMGYY182 pKa = 9.35FEE184 pKa = 5.56IEE186 pKa = 4.17TNKK189 pKa = 10.34EE190 pKa = 3.9LTFGTLIHH198 pKa = 6.65FLRR201 pKa = 11.84EE202 pKa = 3.78KK203 pKa = 11.13YY204 pKa = 10.64SFNKK208 pKa = 10.25LFIII212 pKa = 5.22
Molecular weight: 24.78 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1W5W6D7|A0A1W5W6D7_9VIRU Coat protein OS=Watermelon virus A OX=1978413 PE=4 SV=1
MM1 pKa = 7.82ASTSMTLVKK10 pKa = 10.53SLEE13 pKa = 3.9KK14 pKa = 10.32RR15 pKa = 11.84IEE17 pKa = 4.06ADD19 pKa = 3.01KK20 pKa = 11.62SLMNSSEE27 pKa = 3.77VDD29 pKa = 3.04KK30 pKa = 11.28LYY32 pKa = 11.42GNLATHH38 pKa = 5.95VFKK41 pKa = 11.34DD42 pKa = 3.64EE43 pKa = 4.1VKK45 pKa = 10.49IKK47 pKa = 10.57IGGNEE52 pKa = 3.91NGNVVSTQCRR62 pKa = 11.84LIGASRR68 pKa = 11.84MQQILNSAQGKK79 pKa = 8.11KK80 pKa = 9.82AAYY83 pKa = 9.56LHH85 pKa = 6.71IGCIPITIQSLLPSGNSKK103 pKa = 10.31IKK105 pKa = 10.29GRR107 pKa = 11.84CALVDD112 pKa = 3.5VSRR115 pKa = 11.84TSLDD119 pKa = 2.94TGLIDD124 pKa = 4.57GFTFSFTDD132 pKa = 3.39EE133 pKa = 4.05KK134 pKa = 10.71PYY136 pKa = 8.56TGRR139 pKa = 11.84MIIVNKK145 pKa = 10.24VIDD148 pKa = 4.22FEE150 pKa = 5.86CDD152 pKa = 2.96ASVGSIQLVLEE163 pKa = 4.25VEE165 pKa = 4.84GIDD168 pKa = 5.37LRR170 pKa = 11.84IQRR173 pKa = 11.84SALAVTVGMTCIPSNTAIVFPSLQRR198 pKa = 11.84SKK200 pKa = 8.34MTWALTNSVDD210 pKa = 4.4MKK212 pKa = 10.01EE213 pKa = 4.08QEE215 pKa = 4.16IKK217 pKa = 10.53EE218 pKa = 4.07DD219 pKa = 3.65EE220 pKa = 4.16QFKK223 pKa = 10.99EE224 pKa = 4.2LFEE227 pKa = 4.24NTNTNLIDD235 pKa = 3.41QGEE238 pKa = 4.46TVLLEE243 pKa = 4.58EE244 pKa = 5.0KK245 pKa = 10.91GSMLNLFGTKK255 pKa = 9.17RR256 pKa = 11.84KK257 pKa = 7.41TVARR261 pKa = 11.84RR262 pKa = 11.84ILKK265 pKa = 10.3NKK267 pKa = 10.09GPICRR272 pKa = 11.84IEE274 pKa = 3.83NWDD277 pKa = 3.51MGSVKK282 pKa = 10.43SGPVKK287 pKa = 10.2MKK289 pKa = 10.36RR290 pKa = 11.84SNSLSSASSFEE301 pKa = 4.35SKK303 pKa = 10.53FDD305 pKa = 3.45GRR307 pKa = 11.84FARR310 pKa = 11.84QSTSGHH316 pKa = 5.08SRR318 pKa = 11.84EE319 pKa = 4.28TSSS322 pKa = 3.91
MM1 pKa = 7.82ASTSMTLVKK10 pKa = 10.53SLEE13 pKa = 3.9KK14 pKa = 10.32RR15 pKa = 11.84IEE17 pKa = 4.06ADD19 pKa = 3.01KK20 pKa = 11.62SLMNSSEE27 pKa = 3.77VDD29 pKa = 3.04KK30 pKa = 11.28LYY32 pKa = 11.42GNLATHH38 pKa = 5.95VFKK41 pKa = 11.34DD42 pKa = 3.64EE43 pKa = 4.1VKK45 pKa = 10.49IKK47 pKa = 10.57IGGNEE52 pKa = 3.91NGNVVSTQCRR62 pKa = 11.84LIGASRR68 pKa = 11.84MQQILNSAQGKK79 pKa = 8.11KK80 pKa = 9.82AAYY83 pKa = 9.56LHH85 pKa = 6.71IGCIPITIQSLLPSGNSKK103 pKa = 10.31IKK105 pKa = 10.29GRR107 pKa = 11.84CALVDD112 pKa = 3.5VSRR115 pKa = 11.84TSLDD119 pKa = 2.94TGLIDD124 pKa = 4.57GFTFSFTDD132 pKa = 3.39EE133 pKa = 4.05KK134 pKa = 10.71PYY136 pKa = 8.56TGRR139 pKa = 11.84MIIVNKK145 pKa = 10.24VIDD148 pKa = 4.22FEE150 pKa = 5.86CDD152 pKa = 2.96ASVGSIQLVLEE163 pKa = 4.25VEE165 pKa = 4.84GIDD168 pKa = 5.37LRR170 pKa = 11.84IQRR173 pKa = 11.84SALAVTVGMTCIPSNTAIVFPSLQRR198 pKa = 11.84SKK200 pKa = 8.34MTWALTNSVDD210 pKa = 4.4MKK212 pKa = 10.01EE213 pKa = 4.08QEE215 pKa = 4.16IKK217 pKa = 10.53EE218 pKa = 4.07DD219 pKa = 3.65EE220 pKa = 4.16QFKK223 pKa = 10.99EE224 pKa = 4.2LFEE227 pKa = 4.24NTNTNLIDD235 pKa = 3.41QGEE238 pKa = 4.46TVLLEE243 pKa = 4.58EE244 pKa = 5.0KK245 pKa = 10.91GSMLNLFGTKK255 pKa = 9.17RR256 pKa = 11.84KK257 pKa = 7.41TVARR261 pKa = 11.84RR262 pKa = 11.84ILKK265 pKa = 10.3NKK267 pKa = 10.09GPICRR272 pKa = 11.84IEE274 pKa = 3.83NWDD277 pKa = 3.51MGSVKK282 pKa = 10.43SGPVKK287 pKa = 10.2MKK289 pKa = 10.36RR290 pKa = 11.84SNSLSSASSFEE301 pKa = 4.35SKK303 pKa = 10.53FDD305 pKa = 3.45GRR307 pKa = 11.84FARR310 pKa = 11.84QSTSGHH316 pKa = 5.08SRR318 pKa = 11.84EE319 pKa = 4.28TSSS322 pKa = 3.91
Molecular weight: 35.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2598 |
212 |
1826 |
649.5 |
74.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.081 ± 0.929 | 1.694 ± 0.104 |
5.504 ± 0.251 | 9.584 ± 1.075 |
6.12 ± 0.798 | 5.312 ± 0.567 |
2.194 ± 0.338 | 6.351 ± 0.615 |
9.623 ± 0.417 | 9.045 ± 1.277 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.272 ± 0.194 | 5.581 ± 0.547 |
2.348 ± 0.343 | 2.656 ± 0.385 |
5.119 ± 0.291 | 7.121 ± 1.192 |
4.311 ± 0.769 | 5.504 ± 0.34 |
0.962 ± 0.181 | 2.617 ± 0.566 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
