
Eubacterium sp. CAG:202
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Eubacteriaceae; Eubacterium; environmental samples
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins
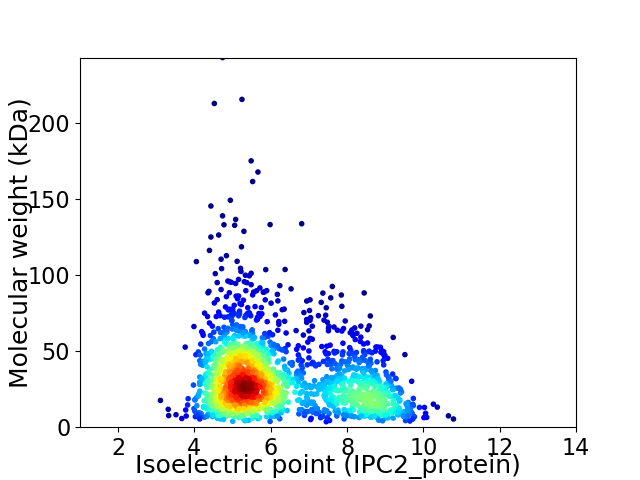
Virtual 2D-PAGE plot for 1817 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R6MTT3|R6MTT3_9FIRM Pseudouridine synthase OS=Eubacterium sp. CAG:202 OX=1262884 GN=BN531_01333 PE=3 SV=1
MM1 pKa = 7.19KK2 pKa = 8.24TTAKK6 pKa = 10.31FISASLATLLVSASAVTAFAAEE28 pKa = 4.38TKK30 pKa = 10.42DD31 pKa = 3.03ITVSLRR37 pKa = 11.84IEE39 pKa = 4.45GVDD42 pKa = 3.69SCVLYY47 pKa = 10.86DD48 pKa = 4.4NYY50 pKa = 10.55EE51 pKa = 3.93IPQGSTAADD60 pKa = 4.9LIQYY64 pKa = 10.16ADD66 pKa = 4.05KK67 pKa = 11.21LSDD70 pKa = 3.6DD71 pKa = 3.9VTVTGAEE78 pKa = 3.87NNYY81 pKa = 7.99ITDD84 pKa = 3.7VNGEE88 pKa = 4.13TAGKK92 pKa = 9.98FGGWDD97 pKa = 2.85GWQYY101 pKa = 11.15IVNSVSPNVGVGDD114 pKa = 3.86YY115 pKa = 9.69TLSDD119 pKa = 3.58NDD121 pKa = 4.23TVVLYY126 pKa = 11.13YY127 pKa = 11.13GDD129 pKa = 4.77FPCLLPQIDD138 pKa = 3.94TSALNSDD145 pKa = 3.78GKK147 pKa = 10.8ISFNAEE153 pKa = 3.61STTYY157 pKa = 11.09DD158 pKa = 3.5SDD160 pKa = 3.69WNPTVSTVAIADD172 pKa = 3.55MTVTFDD178 pKa = 2.87GHH180 pKa = 6.6TYY182 pKa = 7.78TTDD185 pKa = 3.23EE186 pKa = 4.69NGTISLSKK194 pKa = 10.92ADD196 pKa = 3.92FTSGEE201 pKa = 4.02HH202 pKa = 5.6SVQVEE207 pKa = 4.26KK208 pKa = 11.04KK209 pKa = 9.61NSNGAPAVLRR219 pKa = 11.84YY220 pKa = 10.11ADD222 pKa = 4.48DD223 pKa = 3.63FTVNIARR230 pKa = 11.84EE231 pKa = 3.78NTINVNLRR239 pKa = 11.84IEE241 pKa = 4.43GPEE244 pKa = 3.61ACYY247 pKa = 10.64LKK249 pKa = 10.24DD250 pKa = 3.18TFEE253 pKa = 4.09IAKK256 pKa = 10.04DD257 pKa = 3.76SNVGQLISYY266 pKa = 10.19ADD268 pKa = 3.64EE269 pKa = 4.12VSDD272 pKa = 4.09NIEE275 pKa = 4.08VVGADD280 pKa = 2.8AGYY283 pKa = 9.26ISEE286 pKa = 4.56VNGIAAGSFGGWDD299 pKa = 2.95GWYY302 pKa = 9.47YY303 pKa = 11.28AVNSVVPNVGVNDD316 pKa = 3.55YY317 pKa = 9.04TLNNNDD323 pKa = 3.77TVVLYY328 pKa = 10.73YY329 pKa = 10.87GEE331 pKa = 4.42YY332 pKa = 9.83PCYY335 pKa = 10.53LPIADD340 pKa = 4.18TSLLTSEE347 pKa = 4.71GKK349 pKa = 8.14ITFTATATFGDD360 pKa = 4.08AVLPIDD366 pKa = 3.96NMTVYY371 pKa = 10.59FDD373 pKa = 3.54GKK375 pKa = 10.92EE376 pKa = 3.79YY377 pKa = 8.12TTDD380 pKa = 3.34YY381 pKa = 10.87NGVVVIDD388 pKa = 4.06KK389 pKa = 10.63DD390 pKa = 3.58QLTFGSHH397 pKa = 5.15SLQVEE402 pKa = 4.41KK403 pKa = 10.83YY404 pKa = 8.86GYY406 pKa = 9.91SGAPAVLRR414 pKa = 11.84YY415 pKa = 10.1ADD417 pKa = 4.72DD418 pKa = 3.59YY419 pKa = 11.64TVFVDD424 pKa = 3.77TKK426 pKa = 10.57IVNDD430 pKa = 3.48VNLDD434 pKa = 3.72GNVDD438 pKa = 3.62INDD441 pKa = 3.21VTAIQTYY448 pKa = 9.89LSNYY452 pKa = 9.16NNISEE457 pKa = 4.33EE458 pKa = 3.97QVRR461 pKa = 11.84IADD464 pKa = 3.59VNKK467 pKa = 10.51DD468 pKa = 3.21GKK470 pKa = 11.27VDD472 pKa = 3.91VNDD475 pKa = 3.31VTALQTILSGEE486 pKa = 4.31TEE488 pKa = 3.91
MM1 pKa = 7.19KK2 pKa = 8.24TTAKK6 pKa = 10.31FISASLATLLVSASAVTAFAAEE28 pKa = 4.38TKK30 pKa = 10.42DD31 pKa = 3.03ITVSLRR37 pKa = 11.84IEE39 pKa = 4.45GVDD42 pKa = 3.69SCVLYY47 pKa = 10.86DD48 pKa = 4.4NYY50 pKa = 10.55EE51 pKa = 3.93IPQGSTAADD60 pKa = 4.9LIQYY64 pKa = 10.16ADD66 pKa = 4.05KK67 pKa = 11.21LSDD70 pKa = 3.6DD71 pKa = 3.9VTVTGAEE78 pKa = 3.87NNYY81 pKa = 7.99ITDD84 pKa = 3.7VNGEE88 pKa = 4.13TAGKK92 pKa = 9.98FGGWDD97 pKa = 2.85GWQYY101 pKa = 11.15IVNSVSPNVGVGDD114 pKa = 3.86YY115 pKa = 9.69TLSDD119 pKa = 3.58NDD121 pKa = 4.23TVVLYY126 pKa = 11.13YY127 pKa = 11.13GDD129 pKa = 4.77FPCLLPQIDD138 pKa = 3.94TSALNSDD145 pKa = 3.78GKK147 pKa = 10.8ISFNAEE153 pKa = 3.61STTYY157 pKa = 11.09DD158 pKa = 3.5SDD160 pKa = 3.69WNPTVSTVAIADD172 pKa = 3.55MTVTFDD178 pKa = 2.87GHH180 pKa = 6.6TYY182 pKa = 7.78TTDD185 pKa = 3.23EE186 pKa = 4.69NGTISLSKK194 pKa = 10.92ADD196 pKa = 3.92FTSGEE201 pKa = 4.02HH202 pKa = 5.6SVQVEE207 pKa = 4.26KK208 pKa = 11.04KK209 pKa = 9.61NSNGAPAVLRR219 pKa = 11.84YY220 pKa = 10.11ADD222 pKa = 4.48DD223 pKa = 3.63FTVNIARR230 pKa = 11.84EE231 pKa = 3.78NTINVNLRR239 pKa = 11.84IEE241 pKa = 4.43GPEE244 pKa = 3.61ACYY247 pKa = 10.64LKK249 pKa = 10.24DD250 pKa = 3.18TFEE253 pKa = 4.09IAKK256 pKa = 10.04DD257 pKa = 3.76SNVGQLISYY266 pKa = 10.19ADD268 pKa = 3.64EE269 pKa = 4.12VSDD272 pKa = 4.09NIEE275 pKa = 4.08VVGADD280 pKa = 2.8AGYY283 pKa = 9.26ISEE286 pKa = 4.56VNGIAAGSFGGWDD299 pKa = 2.95GWYY302 pKa = 9.47YY303 pKa = 11.28AVNSVVPNVGVNDD316 pKa = 3.55YY317 pKa = 9.04TLNNNDD323 pKa = 3.77TVVLYY328 pKa = 10.73YY329 pKa = 10.87GEE331 pKa = 4.42YY332 pKa = 9.83PCYY335 pKa = 10.53LPIADD340 pKa = 4.18TSLLTSEE347 pKa = 4.71GKK349 pKa = 8.14ITFTATATFGDD360 pKa = 4.08AVLPIDD366 pKa = 3.96NMTVYY371 pKa = 10.59FDD373 pKa = 3.54GKK375 pKa = 10.92EE376 pKa = 3.79YY377 pKa = 8.12TTDD380 pKa = 3.34YY381 pKa = 10.87NGVVVIDD388 pKa = 4.06KK389 pKa = 10.63DD390 pKa = 3.58QLTFGSHH397 pKa = 5.15SLQVEE402 pKa = 4.41KK403 pKa = 10.83YY404 pKa = 8.86GYY406 pKa = 9.91SGAPAVLRR414 pKa = 11.84YY415 pKa = 10.1ADD417 pKa = 4.72DD418 pKa = 3.59YY419 pKa = 11.64TVFVDD424 pKa = 3.77TKK426 pKa = 10.57IVNDD430 pKa = 3.48VNLDD434 pKa = 3.72GNVDD438 pKa = 3.62INDD441 pKa = 3.21VTAIQTYY448 pKa = 9.89LSNYY452 pKa = 9.16NNISEE457 pKa = 4.33EE458 pKa = 3.97QVRR461 pKa = 11.84IADD464 pKa = 3.59VNKK467 pKa = 10.51DD468 pKa = 3.21GKK470 pKa = 11.27VDD472 pKa = 3.91VNDD475 pKa = 3.31VTALQTILSGEE486 pKa = 4.31TEE488 pKa = 3.91
Molecular weight: 52.72 kDa
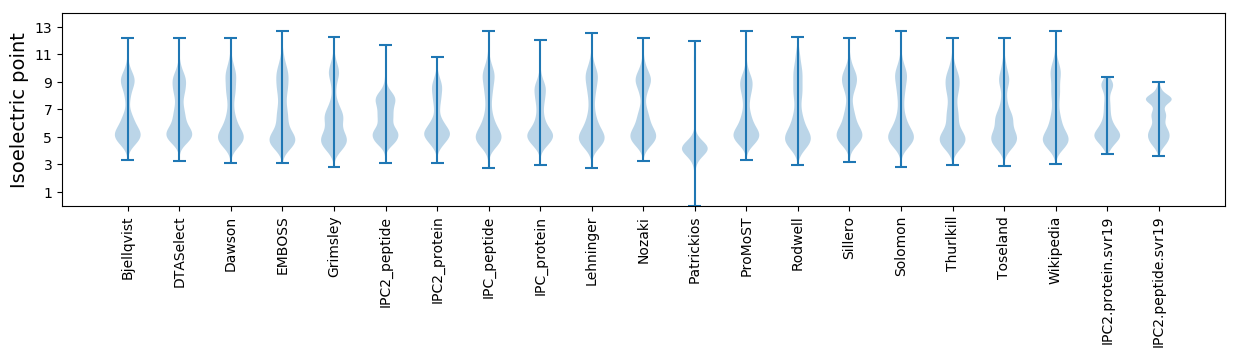
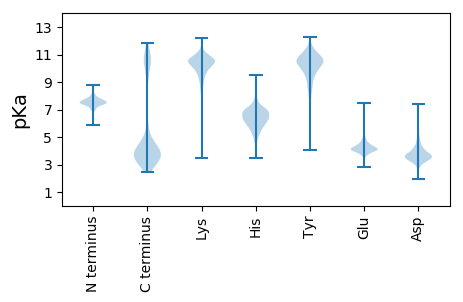
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R6N451|R6N451_9FIRM Uncharacterized protein OS=Eubacterium sp. CAG:202 OX=1262884 GN=BN531_00499 PE=3 SV=1
MM1 pKa = 8.11ADD3 pKa = 3.49RR4 pKa = 11.84PINRR8 pKa = 11.84GRR10 pKa = 11.84KK11 pKa = 8.61ARR13 pKa = 11.84KK14 pKa = 8.66KK15 pKa = 9.55VCGFCVDD22 pKa = 3.51KK23 pKa = 11.43VEE25 pKa = 5.09HH26 pKa = 6.88IDD28 pKa = 3.57YY29 pKa = 10.97KK30 pKa = 11.13DD31 pKa = 2.98IARR34 pKa = 11.84LRR36 pKa = 11.84RR37 pKa = 11.84YY38 pKa = 8.42MSEE41 pKa = 3.7RR42 pKa = 11.84GKK44 pKa = 10.14ILPRR48 pKa = 11.84RR49 pKa = 11.84VTGTCAKK56 pKa = 8.92HH57 pKa = 4.75QRR59 pKa = 11.84EE60 pKa = 3.87ITVAIKK66 pKa = 10.38RR67 pKa = 11.84ARR69 pKa = 11.84HH70 pKa = 5.37LALLPYY76 pKa = 9.54TADD79 pKa = 3.2
MM1 pKa = 8.11ADD3 pKa = 3.49RR4 pKa = 11.84PINRR8 pKa = 11.84GRR10 pKa = 11.84KK11 pKa = 8.61ARR13 pKa = 11.84KK14 pKa = 8.66KK15 pKa = 9.55VCGFCVDD22 pKa = 3.51KK23 pKa = 11.43VEE25 pKa = 5.09HH26 pKa = 6.88IDD28 pKa = 3.57YY29 pKa = 10.97KK30 pKa = 11.13DD31 pKa = 2.98IARR34 pKa = 11.84LRR36 pKa = 11.84RR37 pKa = 11.84YY38 pKa = 8.42MSEE41 pKa = 3.7RR42 pKa = 11.84GKK44 pKa = 10.14ILPRR48 pKa = 11.84RR49 pKa = 11.84VTGTCAKK56 pKa = 8.92HH57 pKa = 4.75QRR59 pKa = 11.84EE60 pKa = 3.87ITVAIKK66 pKa = 10.38RR67 pKa = 11.84ARR69 pKa = 11.84HH70 pKa = 5.37LALLPYY76 pKa = 9.54TADD79 pKa = 3.2
Molecular weight: 9.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
547490 |
32 |
2146 |
301.3 |
33.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.351 ± 0.062 | 1.685 ± 0.024 |
5.99 ± 0.046 | 6.673 ± 0.058 |
4.412 ± 0.045 | 6.642 ± 0.05 |
1.426 ± 0.026 | 7.933 ± 0.058 |
7.727 ± 0.052 | 8.316 ± 0.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.672 ± 0.027 | 5.606 ± 0.054 |
3.14 ± 0.034 | 2.633 ± 0.029 |
3.541 ± 0.046 | 6.51 ± 0.055 |
5.812 ± 0.055 | 7.032 ± 0.049 |
0.707 ± 0.018 | 4.192 ± 0.045 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
