
Datura yellow vein nucleorhabdovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Nucleorhabdovirus
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins
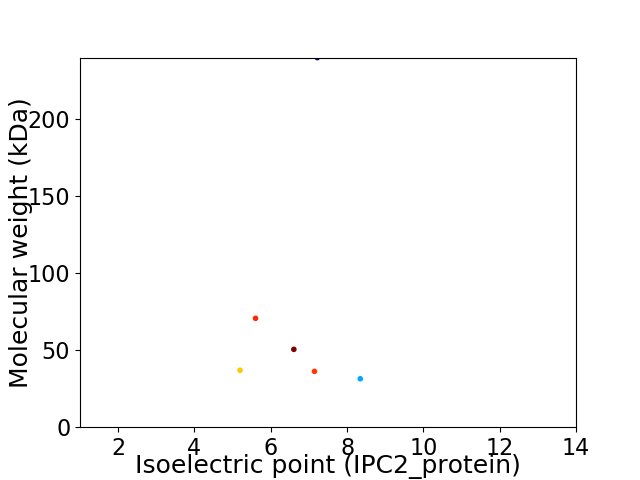
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0F7LI32|A0A0F7LI32_9RHAB Phosphoprotein OS=Datura yellow vein nucleorhabdovirus OX=195059 GN=P PE=4 SV=1
MM1 pKa = 7.6SDD3 pKa = 3.33KK4 pKa = 10.78VHH6 pKa = 6.76PRR8 pKa = 11.84YY9 pKa = 9.83QSLPKK14 pKa = 9.47TATSSEE20 pKa = 4.08VMASKK25 pKa = 10.58YY26 pKa = 10.09AAEE29 pKa = 4.06ISKK32 pKa = 9.03TGNTEE37 pKa = 3.76DD38 pKa = 4.32DD39 pKa = 4.2EE40 pKa = 5.82EE41 pKa = 4.28MMKK44 pKa = 9.54YY45 pKa = 8.08TEE47 pKa = 3.83EE48 pKa = 4.18WEE50 pKa = 3.99EE51 pKa = 3.79HH52 pKa = 5.0FRR54 pKa = 11.84RR55 pKa = 11.84EE56 pKa = 3.98GLTISNRR63 pKa = 11.84LIATLSEE70 pKa = 4.05VTLLCKK76 pKa = 10.45KK77 pKa = 10.24NNKK80 pKa = 9.25EE81 pKa = 3.77EE82 pKa = 4.16LAGKK86 pKa = 9.58LATTLVDD93 pKa = 3.53SFQSVIRR100 pKa = 11.84QHH102 pKa = 5.92TSLSNVVSHH111 pKa = 7.21LEE113 pKa = 3.81ALSEE117 pKa = 4.02NLFTGIDD124 pKa = 3.87EE125 pKa = 4.33IKK127 pKa = 10.87KK128 pKa = 10.61LGTDD132 pKa = 3.59LQKK135 pKa = 11.14ALPRR139 pKa = 11.84KK140 pKa = 9.62KK141 pKa = 10.15ISVSKK146 pKa = 10.55KK147 pKa = 7.56SSRR150 pKa = 11.84PIKK153 pKa = 9.64QAEE156 pKa = 4.32PIYY159 pKa = 10.49DD160 pKa = 3.71LSDD163 pKa = 3.44SPEE166 pKa = 4.05MMKK169 pKa = 10.24HH170 pKa = 4.9QPTTFSIPQEE180 pKa = 4.1QAPGTSSAPPPQTMEE195 pKa = 4.0HH196 pKa = 6.18EE197 pKa = 4.61TTKK200 pKa = 10.78EE201 pKa = 3.79PEE203 pKa = 4.01NNLEE207 pKa = 4.03SEE209 pKa = 4.28AMKK212 pKa = 10.3QLRR215 pKa = 11.84SSYY218 pKa = 9.86RR219 pKa = 11.84GYY221 pKa = 10.28YY222 pKa = 10.35SSDD225 pKa = 3.0EE226 pKa = 4.07FVAFDD231 pKa = 5.1IIKK234 pKa = 9.05QHH236 pKa = 6.12QIVNYY241 pKa = 9.34YY242 pKa = 9.26VEE244 pKa = 4.27NVLGFKK250 pKa = 10.22KK251 pKa = 10.56DD252 pKa = 3.75PEE254 pKa = 4.71DD255 pKa = 4.2KK256 pKa = 10.63DD257 pKa = 3.5PHH259 pKa = 6.54VLSMIFDD266 pKa = 4.13LVDD269 pKa = 3.31KK270 pKa = 10.66EE271 pKa = 4.44RR272 pKa = 11.84VLAVCKK278 pKa = 10.29KK279 pKa = 9.99IKK281 pKa = 9.78TGEE284 pKa = 4.02LDD286 pKa = 3.21EE287 pKa = 5.54ALITDD292 pKa = 3.84SVEE295 pKa = 4.46EE296 pKa = 3.88IVEE299 pKa = 4.75AINKK303 pKa = 8.56CGPAYY308 pKa = 10.26GEE310 pKa = 3.93RR311 pKa = 11.84TAEE314 pKa = 3.95EE315 pKa = 4.6VIVEE319 pKa = 4.09GRR321 pKa = 11.84SRR323 pKa = 11.84LVINN327 pKa = 4.73
MM1 pKa = 7.6SDD3 pKa = 3.33KK4 pKa = 10.78VHH6 pKa = 6.76PRR8 pKa = 11.84YY9 pKa = 9.83QSLPKK14 pKa = 9.47TATSSEE20 pKa = 4.08VMASKK25 pKa = 10.58YY26 pKa = 10.09AAEE29 pKa = 4.06ISKK32 pKa = 9.03TGNTEE37 pKa = 3.76DD38 pKa = 4.32DD39 pKa = 4.2EE40 pKa = 5.82EE41 pKa = 4.28MMKK44 pKa = 9.54YY45 pKa = 8.08TEE47 pKa = 3.83EE48 pKa = 4.18WEE50 pKa = 3.99EE51 pKa = 3.79HH52 pKa = 5.0FRR54 pKa = 11.84RR55 pKa = 11.84EE56 pKa = 3.98GLTISNRR63 pKa = 11.84LIATLSEE70 pKa = 4.05VTLLCKK76 pKa = 10.45KK77 pKa = 10.24NNKK80 pKa = 9.25EE81 pKa = 3.77EE82 pKa = 4.16LAGKK86 pKa = 9.58LATTLVDD93 pKa = 3.53SFQSVIRR100 pKa = 11.84QHH102 pKa = 5.92TSLSNVVSHH111 pKa = 7.21LEE113 pKa = 3.81ALSEE117 pKa = 4.02NLFTGIDD124 pKa = 3.87EE125 pKa = 4.33IKK127 pKa = 10.87KK128 pKa = 10.61LGTDD132 pKa = 3.59LQKK135 pKa = 11.14ALPRR139 pKa = 11.84KK140 pKa = 9.62KK141 pKa = 10.15ISVSKK146 pKa = 10.55KK147 pKa = 7.56SSRR150 pKa = 11.84PIKK153 pKa = 9.64QAEE156 pKa = 4.32PIYY159 pKa = 10.49DD160 pKa = 3.71LSDD163 pKa = 3.44SPEE166 pKa = 4.05MMKK169 pKa = 10.24HH170 pKa = 4.9QPTTFSIPQEE180 pKa = 4.1QAPGTSSAPPPQTMEE195 pKa = 4.0HH196 pKa = 6.18EE197 pKa = 4.61TTKK200 pKa = 10.78EE201 pKa = 3.79PEE203 pKa = 4.01NNLEE207 pKa = 4.03SEE209 pKa = 4.28AMKK212 pKa = 10.3QLRR215 pKa = 11.84SSYY218 pKa = 9.86RR219 pKa = 11.84GYY221 pKa = 10.28YY222 pKa = 10.35SSDD225 pKa = 3.0EE226 pKa = 4.07FVAFDD231 pKa = 5.1IIKK234 pKa = 9.05QHH236 pKa = 6.12QIVNYY241 pKa = 9.34YY242 pKa = 9.26VEE244 pKa = 4.27NVLGFKK250 pKa = 10.22KK251 pKa = 10.56DD252 pKa = 3.75PEE254 pKa = 4.71DD255 pKa = 4.2KK256 pKa = 10.63DD257 pKa = 3.5PHH259 pKa = 6.54VLSMIFDD266 pKa = 4.13LVDD269 pKa = 3.31KK270 pKa = 10.66EE271 pKa = 4.44RR272 pKa = 11.84VLAVCKK278 pKa = 10.29KK279 pKa = 9.99IKK281 pKa = 9.78TGEE284 pKa = 4.02LDD286 pKa = 3.21EE287 pKa = 5.54ALITDD292 pKa = 3.84SVEE295 pKa = 4.46EE296 pKa = 3.88IVEE299 pKa = 4.75AINKK303 pKa = 8.56CGPAYY308 pKa = 10.26GEE310 pKa = 3.93RR311 pKa = 11.84TAEE314 pKa = 3.95EE315 pKa = 4.6VIVEE319 pKa = 4.09GRR321 pKa = 11.84SRR323 pKa = 11.84LVINN327 pKa = 4.73
Molecular weight: 36.92 kDa
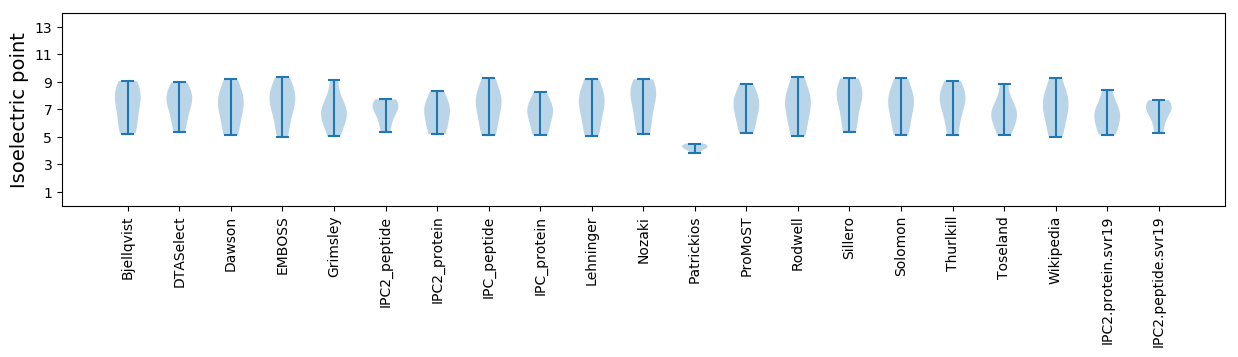
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0F7LG26|A0A0F7LG26_9RHAB Glycoprotein OS=Datura yellow vein nucleorhabdovirus OX=195059 GN=G PE=4 SV=1
MM1 pKa = 7.52EE2 pKa = 5.56PASFSIKK9 pKa = 10.58GNVGLTFKK17 pKa = 10.74KK18 pKa = 10.51KK19 pKa = 9.55RR20 pKa = 11.84DD21 pKa = 3.57SMHH24 pKa = 7.26ALTLWDD30 pKa = 4.1IVTKK34 pKa = 8.86MWNMYY39 pKa = 8.2PEE41 pKa = 4.09RR42 pKa = 11.84VVVSFSTTRR51 pKa = 11.84NSADD55 pKa = 3.28KK56 pKa = 11.27SPVEE60 pKa = 4.38DD61 pKa = 4.17PLVIAAIQKK70 pKa = 8.62MVEE73 pKa = 4.23GCVKK77 pKa = 10.36SYY79 pKa = 11.0NIKK82 pKa = 10.26CSVGRR87 pKa = 11.84SVNLLLGDD95 pKa = 4.05CYY97 pKa = 10.93KK98 pKa = 10.93YY99 pKa = 10.91AIALGTSSKK108 pKa = 10.18DD109 pKa = 3.24TGHH112 pKa = 5.7EE113 pKa = 4.25TIILTLPFSMKK124 pKa = 9.39GCYY127 pKa = 9.26RR128 pKa = 11.84IHH130 pKa = 8.13AEE132 pKa = 4.43VSNHH136 pKa = 4.69MKK138 pKa = 10.32KK139 pKa = 10.58DD140 pKa = 3.66GVKK143 pKa = 8.39EE144 pKa = 3.9HH145 pKa = 6.35YY146 pKa = 9.29MLGIGLEE153 pKa = 4.13AYY155 pKa = 9.5IGDD158 pKa = 4.46MDD160 pKa = 4.23TEE162 pKa = 4.84GYY164 pKa = 9.69NAAIKK169 pKa = 10.24HH170 pKa = 6.44GICIYY175 pKa = 10.44PFMIEE180 pKa = 3.99HH181 pKa = 7.47PEE183 pKa = 4.06LFSKK187 pKa = 10.86SITSDD192 pKa = 3.11SEE194 pKa = 3.96MSSDD198 pKa = 3.16GGHH201 pKa = 5.39YY202 pKa = 10.2VAGPSAVMRR211 pKa = 11.84NKK213 pKa = 9.84KK214 pKa = 9.45RR215 pKa = 11.84RR216 pKa = 11.84KK217 pKa = 9.35QKK219 pKa = 9.94MLLKK223 pKa = 10.2SAKK226 pKa = 9.21PYY228 pKa = 10.39LSGRR232 pKa = 11.84ATSAKK237 pKa = 9.89KK238 pKa = 10.43VLNALRR244 pKa = 11.84LAGNKK249 pKa = 9.28IYY251 pKa = 10.73EE252 pKa = 4.58EE253 pKa = 4.46YY254 pKa = 7.92TTTDD258 pKa = 2.74GDD260 pKa = 3.73EE261 pKa = 4.41AQQQGNNTPSSQEE274 pKa = 3.61EE275 pKa = 4.17KK276 pKa = 11.07APATVPTKK284 pKa = 10.85
MM1 pKa = 7.52EE2 pKa = 5.56PASFSIKK9 pKa = 10.58GNVGLTFKK17 pKa = 10.74KK18 pKa = 10.51KK19 pKa = 9.55RR20 pKa = 11.84DD21 pKa = 3.57SMHH24 pKa = 7.26ALTLWDD30 pKa = 4.1IVTKK34 pKa = 8.86MWNMYY39 pKa = 8.2PEE41 pKa = 4.09RR42 pKa = 11.84VVVSFSTTRR51 pKa = 11.84NSADD55 pKa = 3.28KK56 pKa = 11.27SPVEE60 pKa = 4.38DD61 pKa = 4.17PLVIAAIQKK70 pKa = 8.62MVEE73 pKa = 4.23GCVKK77 pKa = 10.36SYY79 pKa = 11.0NIKK82 pKa = 10.26CSVGRR87 pKa = 11.84SVNLLLGDD95 pKa = 4.05CYY97 pKa = 10.93KK98 pKa = 10.93YY99 pKa = 10.91AIALGTSSKK108 pKa = 10.18DD109 pKa = 3.24TGHH112 pKa = 5.7EE113 pKa = 4.25TIILTLPFSMKK124 pKa = 9.39GCYY127 pKa = 9.26RR128 pKa = 11.84IHH130 pKa = 8.13AEE132 pKa = 4.43VSNHH136 pKa = 4.69MKK138 pKa = 10.32KK139 pKa = 10.58DD140 pKa = 3.66GVKK143 pKa = 8.39EE144 pKa = 3.9HH145 pKa = 6.35YY146 pKa = 9.29MLGIGLEE153 pKa = 4.13AYY155 pKa = 9.5IGDD158 pKa = 4.46MDD160 pKa = 4.23TEE162 pKa = 4.84GYY164 pKa = 9.69NAAIKK169 pKa = 10.24HH170 pKa = 6.44GICIYY175 pKa = 10.44PFMIEE180 pKa = 3.99HH181 pKa = 7.47PEE183 pKa = 4.06LFSKK187 pKa = 10.86SITSDD192 pKa = 3.11SEE194 pKa = 3.96MSSDD198 pKa = 3.16GGHH201 pKa = 5.39YY202 pKa = 10.2VAGPSAVMRR211 pKa = 11.84NKK213 pKa = 9.84KK214 pKa = 9.45RR215 pKa = 11.84RR216 pKa = 11.84KK217 pKa = 9.35QKK219 pKa = 9.94MLLKK223 pKa = 10.2SAKK226 pKa = 9.21PYY228 pKa = 10.39LSGRR232 pKa = 11.84ATSAKK237 pKa = 9.89KK238 pKa = 10.43VLNALRR244 pKa = 11.84LAGNKK249 pKa = 9.28IYY251 pKa = 10.73EE252 pKa = 4.58EE253 pKa = 4.46YY254 pKa = 7.92TTTDD258 pKa = 2.74GDD260 pKa = 3.73EE261 pKa = 4.41AQQQGNNTPSSQEE274 pKa = 3.61EE275 pKa = 4.17KK276 pKa = 11.07APATVPTKK284 pKa = 10.85
Molecular weight: 31.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4118 |
284 |
2106 |
686.3 |
77.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.78 ± 0.969 | 2.064 ± 0.235 |
5.585 ± 0.515 | 6.241 ± 0.771 |
3.084 ± 0.315 | 5.585 ± 0.406 |
2.695 ± 0.191 | 6.848 ± 0.308 |
6.605 ± 0.45 | 8.791 ± 0.787 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.181 ± 0.2 | 4.687 ± 0.142 |
4.177 ± 0.451 | 2.841 ± 0.159 |
5.148 ± 0.446 | 8.451 ± 0.163 |
6.581 ± 0.48 | 6.217 ± 0.335 |
1.36 ± 0.197 | 4.08 ± 0.319 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
