
Idiomarina tyrosinivorans
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Idiomarinaceae; Idiomarina
Average proteome isoelectric point is 6.25
Get precalculated fractions of proteins
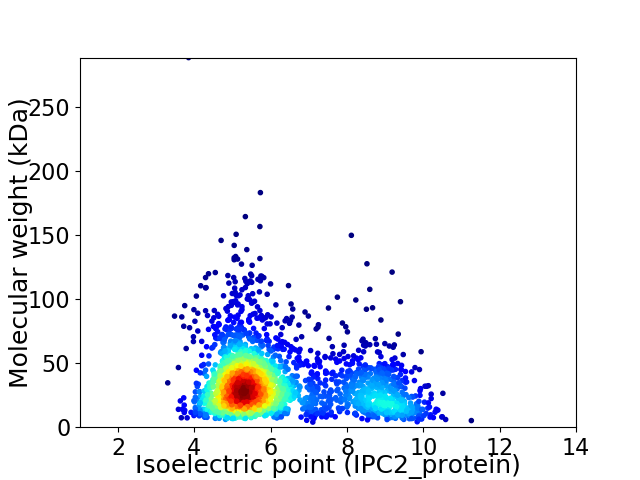
Virtual 2D-PAGE plot for 2274 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A432ZQU4|A0A432ZQU4_9GAMM GTPase Der OS=Idiomarina tyrosinivorans OX=1445662 GN=der PE=3 SV=1
MM1 pKa = 7.56KK2 pKa = 10.1FLHH5 pKa = 6.69SLTAIAVAAALSACGGDD22 pKa = 3.73DD23 pKa = 3.95EE24 pKa = 6.54KK25 pKa = 11.62NVAPIATDD33 pKa = 3.49DD34 pKa = 4.18SYY36 pKa = 11.52TATVGATNTIDD47 pKa = 3.87VLANDD52 pKa = 3.93SDD54 pKa = 4.47EE55 pKa = 4.27NTKK58 pKa = 9.44DD59 pKa = 3.55TLSVTAVTQPDD70 pKa = 3.83SGSVTINSDD79 pKa = 3.03GTLSFEE85 pKa = 4.7PSGDD89 pKa = 3.52TLGDD93 pKa = 3.38DD94 pKa = 3.44SFTYY98 pKa = 9.6TVSDD102 pKa = 4.35GEE104 pKa = 4.45LDD106 pKa = 3.64DD107 pKa = 4.14TATVSLTVQQALAFSGKK124 pKa = 9.89VVDD127 pKa = 4.39AAIPNATVTITIGEE141 pKa = 4.31DD142 pKa = 3.35TFTTTADD149 pKa = 3.28EE150 pKa = 4.78DD151 pKa = 4.06GNYY154 pKa = 8.84TLEE157 pKa = 3.94VFYY160 pKa = 10.86TDD162 pKa = 2.97GTKK165 pKa = 10.11LVKK168 pKa = 10.19IEE170 pKa = 4.25AVGSADD176 pKa = 3.78NEE178 pKa = 4.0QDD180 pKa = 4.08HH181 pKa = 6.92VDD183 pKa = 4.03LTSLMPSLATLQGLAGDD200 pKa = 4.07DD201 pKa = 4.43AIVQRR206 pKa = 11.84SEE208 pKa = 4.18AGGTNVTNVTTASYY222 pKa = 11.52VLTKK226 pKa = 9.87EE227 pKa = 4.33ANGGEE232 pKa = 4.19DD233 pKa = 3.42PADD236 pKa = 3.89EE237 pKa = 4.41NTLSAAEE244 pKa = 4.15SNIDD248 pKa = 3.27GTKK251 pKa = 9.75VLEE254 pKa = 3.88IAAVIKK260 pKa = 10.34VIVDD264 pKa = 3.44NPDD267 pKa = 2.76FDD269 pKa = 5.81LPDD272 pKa = 3.77GVSTIGEE279 pKa = 4.69LINDD283 pKa = 3.32EE284 pKa = 4.27TAYY287 pKa = 10.79NDD289 pKa = 3.14YY290 pKa = 10.73VAYY293 pKa = 10.69VEE295 pKa = 4.43NTDD298 pKa = 3.52PTALQAAADD307 pKa = 4.52AIVEE311 pKa = 4.18DD312 pKa = 4.56EE313 pKa = 4.4EE314 pKa = 4.5LTVASTQFKK323 pKa = 10.81LPAFYY328 pKa = 10.23IQLPTTQPGFIARR341 pKa = 11.84GDD343 pKa = 3.53STLYY347 pKa = 10.98FNDD350 pKa = 4.83DD351 pKa = 3.24GTGVYY356 pKa = 10.3INGFFGLEE364 pKa = 3.97NEE366 pKa = 4.79TDD368 pKa = 4.35FTWSRR373 pKa = 11.84DD374 pKa = 3.61DD375 pKa = 3.53NGYY378 pKa = 9.89YY379 pKa = 10.81ALDD382 pKa = 3.74FATPAVYY389 pKa = 9.45IDD391 pKa = 4.0YY392 pKa = 8.91PAVTDD397 pKa = 3.69EE398 pKa = 4.67TDD400 pKa = 3.29DD401 pKa = 3.74ATILAAYY408 pKa = 9.6DD409 pKa = 3.68DD410 pKa = 4.13ASVYY414 pKa = 9.51QVEE417 pKa = 4.74TEE419 pKa = 4.22VSVSALAFKK428 pKa = 10.94LLVDD432 pKa = 4.77GKK434 pKa = 9.08TVDD437 pKa = 3.62LVQAKK442 pKa = 7.42EE443 pKa = 4.44TYY445 pKa = 8.73TKK447 pKa = 10.52VYY449 pKa = 10.42QPLNGVDD456 pKa = 3.11GKK458 pKa = 9.41TYY460 pKa = 10.1QIPTAQGVRR469 pKa = 11.84KK470 pKa = 8.97FQYY473 pKa = 10.5SYY475 pKa = 11.71LNGDD479 pKa = 4.64ALPKK483 pKa = 10.2IAGTEE488 pKa = 3.84ADD490 pKa = 4.09LTTQQWVLPIVGLAAPGDD508 pKa = 3.76MHH510 pKa = 7.24EE511 pKa = 4.9ALQPDD516 pKa = 5.52LITFNSDD523 pKa = 2.38GSFGSDD529 pKa = 3.31YY530 pKa = 11.4AGVDD534 pKa = 4.23GEE536 pKa = 4.38WSLTDD541 pKa = 4.06SIISMSYY548 pKa = 8.82TSEE551 pKa = 3.79EE552 pKa = 4.1FGDD555 pKa = 3.98VEE557 pKa = 4.29QKK559 pKa = 10.45VQIVSTKK566 pKa = 9.99NGVYY570 pKa = 8.8TAFVEE575 pKa = 4.92VSSEE579 pKa = 3.75AANATWYY586 pKa = 10.45DD587 pKa = 3.28WVAPRR592 pKa = 11.84DD593 pKa = 3.74PEE595 pKa = 4.26YY596 pKa = 10.79TIDD599 pKa = 4.47PADD602 pKa = 4.11LVNTAATMWLTNPNIWYY619 pKa = 10.06ASQWDD624 pKa = 3.72QAEE627 pKa = 4.17NLPSVFTILGWQFDD641 pKa = 4.5EE642 pKa = 4.62AGKK645 pKa = 10.49AYY647 pKa = 7.59RR648 pKa = 11.84TSVGCQEE655 pKa = 4.24YY656 pKa = 10.82YY657 pKa = 10.07IDD659 pKa = 4.15GTCTYY664 pKa = 11.03DD665 pKa = 3.12STLVVLNNSTRR676 pKa = 11.84NYY678 pKa = 10.44RR679 pKa = 11.84VIEE682 pKa = 4.48DD683 pKa = 3.21SSGQQRR689 pKa = 11.84IQMAYY694 pKa = 9.97NFEE697 pKa = 4.34CFGAGNQVDD706 pKa = 4.49ANCAAYY712 pKa = 9.68EE713 pKa = 3.95YY714 pKa = 10.53IPLQEE719 pKa = 4.71AEE721 pKa = 4.36NGQITLLEE729 pKa = 4.32SVFVDD734 pKa = 4.2RR735 pKa = 11.84DD736 pKa = 3.01PSTPVNLVSGFTPRR750 pKa = 11.84VTVSIPFEE758 pKa = 4.03FPQGIEE764 pKa = 3.62NSAFPGATASSGQAMPAGKK783 pKa = 9.77VDD785 pKa = 3.35RR786 pKa = 11.84VSFLGKK792 pKa = 8.62TKK794 pKa = 10.47SAPLVINHH802 pKa = 7.01
MM1 pKa = 7.56KK2 pKa = 10.1FLHH5 pKa = 6.69SLTAIAVAAALSACGGDD22 pKa = 3.73DD23 pKa = 3.95EE24 pKa = 6.54KK25 pKa = 11.62NVAPIATDD33 pKa = 3.49DD34 pKa = 4.18SYY36 pKa = 11.52TATVGATNTIDD47 pKa = 3.87VLANDD52 pKa = 3.93SDD54 pKa = 4.47EE55 pKa = 4.27NTKK58 pKa = 9.44DD59 pKa = 3.55TLSVTAVTQPDD70 pKa = 3.83SGSVTINSDD79 pKa = 3.03GTLSFEE85 pKa = 4.7PSGDD89 pKa = 3.52TLGDD93 pKa = 3.38DD94 pKa = 3.44SFTYY98 pKa = 9.6TVSDD102 pKa = 4.35GEE104 pKa = 4.45LDD106 pKa = 3.64DD107 pKa = 4.14TATVSLTVQQALAFSGKK124 pKa = 9.89VVDD127 pKa = 4.39AAIPNATVTITIGEE141 pKa = 4.31DD142 pKa = 3.35TFTTTADD149 pKa = 3.28EE150 pKa = 4.78DD151 pKa = 4.06GNYY154 pKa = 8.84TLEE157 pKa = 3.94VFYY160 pKa = 10.86TDD162 pKa = 2.97GTKK165 pKa = 10.11LVKK168 pKa = 10.19IEE170 pKa = 4.25AVGSADD176 pKa = 3.78NEE178 pKa = 4.0QDD180 pKa = 4.08HH181 pKa = 6.92VDD183 pKa = 4.03LTSLMPSLATLQGLAGDD200 pKa = 4.07DD201 pKa = 4.43AIVQRR206 pKa = 11.84SEE208 pKa = 4.18AGGTNVTNVTTASYY222 pKa = 11.52VLTKK226 pKa = 9.87EE227 pKa = 4.33ANGGEE232 pKa = 4.19DD233 pKa = 3.42PADD236 pKa = 3.89EE237 pKa = 4.41NTLSAAEE244 pKa = 4.15SNIDD248 pKa = 3.27GTKK251 pKa = 9.75VLEE254 pKa = 3.88IAAVIKK260 pKa = 10.34VIVDD264 pKa = 3.44NPDD267 pKa = 2.76FDD269 pKa = 5.81LPDD272 pKa = 3.77GVSTIGEE279 pKa = 4.69LINDD283 pKa = 3.32EE284 pKa = 4.27TAYY287 pKa = 10.79NDD289 pKa = 3.14YY290 pKa = 10.73VAYY293 pKa = 10.69VEE295 pKa = 4.43NTDD298 pKa = 3.52PTALQAAADD307 pKa = 4.52AIVEE311 pKa = 4.18DD312 pKa = 4.56EE313 pKa = 4.4EE314 pKa = 4.5LTVASTQFKK323 pKa = 10.81LPAFYY328 pKa = 10.23IQLPTTQPGFIARR341 pKa = 11.84GDD343 pKa = 3.53STLYY347 pKa = 10.98FNDD350 pKa = 4.83DD351 pKa = 3.24GTGVYY356 pKa = 10.3INGFFGLEE364 pKa = 3.97NEE366 pKa = 4.79TDD368 pKa = 4.35FTWSRR373 pKa = 11.84DD374 pKa = 3.61DD375 pKa = 3.53NGYY378 pKa = 9.89YY379 pKa = 10.81ALDD382 pKa = 3.74FATPAVYY389 pKa = 9.45IDD391 pKa = 4.0YY392 pKa = 8.91PAVTDD397 pKa = 3.69EE398 pKa = 4.67TDD400 pKa = 3.29DD401 pKa = 3.74ATILAAYY408 pKa = 9.6DD409 pKa = 3.68DD410 pKa = 4.13ASVYY414 pKa = 9.51QVEE417 pKa = 4.74TEE419 pKa = 4.22VSVSALAFKK428 pKa = 10.94LLVDD432 pKa = 4.77GKK434 pKa = 9.08TVDD437 pKa = 3.62LVQAKK442 pKa = 7.42EE443 pKa = 4.44TYY445 pKa = 8.73TKK447 pKa = 10.52VYY449 pKa = 10.42QPLNGVDD456 pKa = 3.11GKK458 pKa = 9.41TYY460 pKa = 10.1QIPTAQGVRR469 pKa = 11.84KK470 pKa = 8.97FQYY473 pKa = 10.5SYY475 pKa = 11.71LNGDD479 pKa = 4.64ALPKK483 pKa = 10.2IAGTEE488 pKa = 3.84ADD490 pKa = 4.09LTTQQWVLPIVGLAAPGDD508 pKa = 3.76MHH510 pKa = 7.24EE511 pKa = 4.9ALQPDD516 pKa = 5.52LITFNSDD523 pKa = 2.38GSFGSDD529 pKa = 3.31YY530 pKa = 11.4AGVDD534 pKa = 4.23GEE536 pKa = 4.38WSLTDD541 pKa = 4.06SIISMSYY548 pKa = 8.82TSEE551 pKa = 3.79EE552 pKa = 4.1FGDD555 pKa = 3.98VEE557 pKa = 4.29QKK559 pKa = 10.45VQIVSTKK566 pKa = 9.99NGVYY570 pKa = 8.8TAFVEE575 pKa = 4.92VSSEE579 pKa = 3.75AANATWYY586 pKa = 10.45DD587 pKa = 3.28WVAPRR592 pKa = 11.84DD593 pKa = 3.74PEE595 pKa = 4.26YY596 pKa = 10.79TIDD599 pKa = 4.47PADD602 pKa = 4.11LVNTAATMWLTNPNIWYY619 pKa = 10.06ASQWDD624 pKa = 3.72QAEE627 pKa = 4.17NLPSVFTILGWQFDD641 pKa = 4.5EE642 pKa = 4.62AGKK645 pKa = 10.49AYY647 pKa = 7.59RR648 pKa = 11.84TSVGCQEE655 pKa = 4.24YY656 pKa = 10.82YY657 pKa = 10.07IDD659 pKa = 4.15GTCTYY664 pKa = 11.03DD665 pKa = 3.12STLVVLNNSTRR676 pKa = 11.84NYY678 pKa = 10.44RR679 pKa = 11.84VIEE682 pKa = 4.48DD683 pKa = 3.21SSGQQRR689 pKa = 11.84IQMAYY694 pKa = 9.97NFEE697 pKa = 4.34CFGAGNQVDD706 pKa = 4.49ANCAAYY712 pKa = 9.68EE713 pKa = 3.95YY714 pKa = 10.53IPLQEE719 pKa = 4.71AEE721 pKa = 4.36NGQITLLEE729 pKa = 4.32SVFVDD734 pKa = 4.2RR735 pKa = 11.84DD736 pKa = 3.01PSTPVNLVSGFTPRR750 pKa = 11.84VTVSIPFEE758 pKa = 4.03FPQGIEE764 pKa = 3.62NSAFPGATASSGQAMPAGKK783 pKa = 9.77VDD785 pKa = 3.35RR786 pKa = 11.84VSFLGKK792 pKa = 8.62TKK794 pKa = 10.47SAPLVINHH802 pKa = 7.01
Molecular weight: 85.99 kDa
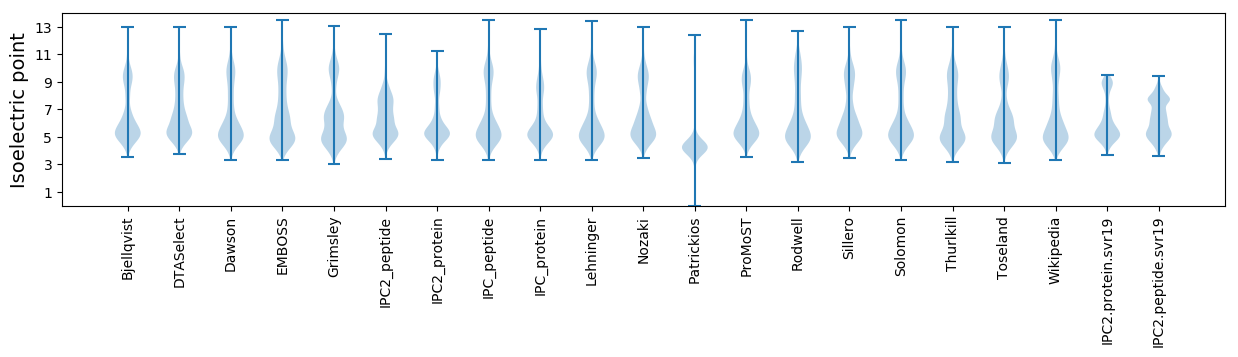
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A432ZTA2|A0A432ZTA2_9GAMM Peptidase M23 OS=Idiomarina tyrosinivorans OX=1445662 GN=CWI84_03185 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.42RR12 pKa = 11.84KK13 pKa = 9.36RR14 pKa = 11.84SHH16 pKa = 6.15GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37NGRR28 pKa = 11.84KK29 pKa = 8.96VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.91VLSAA44 pKa = 4.11
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.42RR12 pKa = 11.84KK13 pKa = 9.36RR14 pKa = 11.84SHH16 pKa = 6.15GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37NGRR28 pKa = 11.84KK29 pKa = 8.96VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.91VLSAA44 pKa = 4.11
Molecular weight: 5.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
746298 |
35 |
2761 |
328.2 |
36.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.859 ± 0.054 | 0.882 ± 0.017 |
5.667 ± 0.048 | 6.125 ± 0.052 |
3.757 ± 0.034 | 6.863 ± 0.054 |
2.258 ± 0.029 | 5.578 ± 0.036 |
4.367 ± 0.045 | 10.556 ± 0.061 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.396 ± 0.027 | 3.645 ± 0.041 |
4.134 ± 0.035 | 5.854 ± 0.059 |
5.597 ± 0.04 | 5.987 ± 0.046 |
5.046 ± 0.038 | 7.11 ± 0.045 |
1.42 ± 0.025 | 2.897 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
