
Meyerozyma guilliermondii (strain ATCC 6260 / CBS 566 / DSM 6381 / JCM 1539 / NBRC 10279 / NRRL Y-324) (Yeast) (Candida guilliermondii)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Saccharomycotina; Saccharomycetes; Saccharomycetales; Debaryomycetaceae; Meyerozyma; Meyerozyma guilliermondii
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
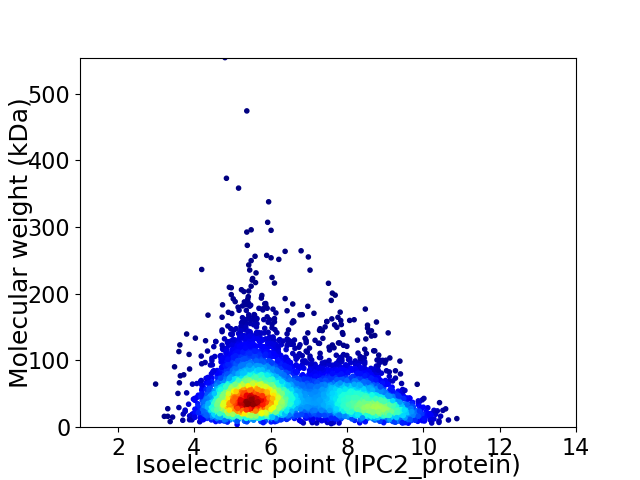
Virtual 2D-PAGE plot for 5919 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A5DI00|A5DI00_PICGU Uncharacterized protein OS=Meyerozyma guilliermondii (strain ATCC 6260 / CBS 566 / DSM 6381 / JCM 1539 / NBRC 10279 / NRRL Y-324) OX=294746 GN=PGUG_02901 PE=4 SV=2
MM1 pKa = 7.76KK2 pKa = 9.26ITTIFHH8 pKa = 5.87TVLLASAAVNAQDD21 pKa = 3.59APKK24 pKa = 10.51KK25 pKa = 10.44RR26 pKa = 11.84GFLSDD31 pKa = 4.06LFGGSDD37 pKa = 3.33AAAPAAAPAAASAATSSTTAAAAAAPVAAATSSAAGVLDD76 pKa = 4.85LFGLDD81 pKa = 3.63SSNSNSTSSSSSSSSSSSSSSSSSSSSSSSSSTKK115 pKa = 10.3APAFRR120 pKa = 11.84VLAAQGTTQSSSSAPATTDD139 pKa = 2.87SGSLFDD145 pKa = 5.19FLNVDD150 pKa = 4.02SSASSSSSSPASSSSKK166 pKa = 10.2VSSSSQTGGLLNQLFAVAPSGSSGSSGSSSASASASKK203 pKa = 10.5SASASSASASVTAAPLGFLGDD224 pKa = 4.04LFGGSSSSKK233 pKa = 9.91ASSGSVSGSSSASSADD249 pKa = 4.36DD250 pKa = 4.07EE251 pKa = 4.67EE252 pKa = 6.7CDD254 pKa = 5.43DD255 pKa = 4.39EE256 pKa = 4.67TDD258 pKa = 3.89SASPTDD264 pKa = 4.45DD265 pKa = 3.56PVSVPPAFSSMSLPPLTGSPSGSLTSLAAVSGQPTSGASRR305 pKa = 11.84SSGVSSARR313 pKa = 11.84SLAASARR320 pKa = 11.84SSVSASSEE328 pKa = 3.86EE329 pKa = 4.09DD330 pKa = 4.3DD331 pKa = 4.32EE332 pKa = 5.56DD333 pKa = 5.89CEE335 pKa = 4.73TEE337 pKa = 4.16SSLPTLPPLPSLSGSGGVLPTLSGSGGILPTLSGSGGILPTLGSASRR384 pKa = 11.84NSTGSTSATVSRR396 pKa = 11.84STASDD401 pKa = 3.64DD402 pKa = 5.68GDD404 pKa = 6.12DD405 pKa = 4.14EE406 pKa = 6.26DD407 pKa = 5.75CDD409 pKa = 4.05PSEE412 pKa = 4.24TTIKK416 pKa = 10.76GDD418 pKa = 3.29PTYY421 pKa = 10.56ATLPTEE427 pKa = 4.03ITSNGDD433 pKa = 3.0PTGITTRR440 pKa = 11.84GTATTTVITTVKK452 pKa = 6.89TTNGGSAVTYY462 pKa = 6.96TTTKK466 pKa = 10.1VVQPTQGSEE475 pKa = 3.85DD476 pKa = 4.35DD477 pKa = 4.16EE478 pKa = 5.35DD479 pKa = 4.88CEE481 pKa = 4.37EE482 pKa = 5.48YY483 pKa = 9.62YY484 pKa = 9.96TQTQTYY490 pKa = 6.87YY491 pKa = 7.96TTYY494 pKa = 7.66TTVINGATVTKK505 pKa = 10.51VVEE508 pKa = 4.12CDD510 pKa = 3.03EE511 pKa = 4.57CKK513 pKa = 10.35GTGSGSGSSGSGSSGSGSSGSGSYY537 pKa = 10.82GGGSSSGSGVDD548 pKa = 3.98EE549 pKa = 4.72DD550 pKa = 5.64CEE552 pKa = 4.68DD553 pKa = 4.62DD554 pKa = 4.19EE555 pKa = 5.6SSPSYY560 pKa = 11.14GSGSGSGTSYY570 pKa = 11.44GSGSGSSGSGSNYY583 pKa = 9.89GSGSGSSSGSGSGSGSTSKK602 pKa = 10.38PGYY605 pKa = 9.74LIPSDD610 pKa = 4.08DD611 pKa = 4.06QCDD614 pKa = 3.92EE615 pKa = 4.4DD616 pKa = 6.2CVVCDD621 pKa = 5.59DD622 pKa = 5.63SDD624 pKa = 6.34DD625 pKa = 4.87EE626 pKa = 4.96DD627 pKa = 5.29FDD629 pKa = 4.07WFGNDD634 pKa = 4.16TDD636 pKa = 4.9DD637 pKa = 5.39EE638 pKa = 5.37CDD640 pKa = 3.85ADD642 pKa = 5.54CDD644 pKa = 3.77EE645 pKa = 5.33CDD647 pKa = 3.68AGTSGSTSGSTSGSGSGSGSGSGSGSGSKK676 pKa = 10.14SGSGSGSGSGGSGSGSGSGGSGGAKK701 pKa = 9.26TYY703 pKa = 10.84VSTVRR708 pKa = 11.84TTAAGEE714 pKa = 4.37STWYY718 pKa = 8.1STSVSTSTHH727 pKa = 6.05SPDD730 pKa = 5.14DD731 pKa = 3.66EE732 pKa = 6.58CDD734 pKa = 3.76DD735 pKa = 4.42EE736 pKa = 5.62EE737 pKa = 5.18SATTTKK743 pKa = 10.73SATTTSGYY751 pKa = 11.39ANSTSKK757 pKa = 8.68QTGGFLADD765 pKa = 3.74LFGRR769 pKa = 11.84DD770 pKa = 3.72VEE772 pKa = 4.48ISSSAWANTSVTSAPVVEE790 pKa = 4.16QHH792 pKa = 6.8PSNGSRR798 pKa = 11.84QTSFSTALVALVVSIVAFTLII819 pKa = 4.01
MM1 pKa = 7.76KK2 pKa = 9.26ITTIFHH8 pKa = 5.87TVLLASAAVNAQDD21 pKa = 3.59APKK24 pKa = 10.51KK25 pKa = 10.44RR26 pKa = 11.84GFLSDD31 pKa = 4.06LFGGSDD37 pKa = 3.33AAAPAAAPAAASAATSSTTAAAAAAPVAAATSSAAGVLDD76 pKa = 4.85LFGLDD81 pKa = 3.63SSNSNSTSSSSSSSSSSSSSSSSSSSSSSSSSTKK115 pKa = 10.3APAFRR120 pKa = 11.84VLAAQGTTQSSSSAPATTDD139 pKa = 2.87SGSLFDD145 pKa = 5.19FLNVDD150 pKa = 4.02SSASSSSSSPASSSSKK166 pKa = 10.2VSSSSQTGGLLNQLFAVAPSGSSGSSGSSSASASASKK203 pKa = 10.5SASASSASASVTAAPLGFLGDD224 pKa = 4.04LFGGSSSSKK233 pKa = 9.91ASSGSVSGSSSASSADD249 pKa = 4.36DD250 pKa = 4.07EE251 pKa = 4.67EE252 pKa = 6.7CDD254 pKa = 5.43DD255 pKa = 4.39EE256 pKa = 4.67TDD258 pKa = 3.89SASPTDD264 pKa = 4.45DD265 pKa = 3.56PVSVPPAFSSMSLPPLTGSPSGSLTSLAAVSGQPTSGASRR305 pKa = 11.84SSGVSSARR313 pKa = 11.84SLAASARR320 pKa = 11.84SSVSASSEE328 pKa = 3.86EE329 pKa = 4.09DD330 pKa = 4.3DD331 pKa = 4.32EE332 pKa = 5.56DD333 pKa = 5.89CEE335 pKa = 4.73TEE337 pKa = 4.16SSLPTLPPLPSLSGSGGVLPTLSGSGGILPTLSGSGGILPTLGSASRR384 pKa = 11.84NSTGSTSATVSRR396 pKa = 11.84STASDD401 pKa = 3.64DD402 pKa = 5.68GDD404 pKa = 6.12DD405 pKa = 4.14EE406 pKa = 6.26DD407 pKa = 5.75CDD409 pKa = 4.05PSEE412 pKa = 4.24TTIKK416 pKa = 10.76GDD418 pKa = 3.29PTYY421 pKa = 10.56ATLPTEE427 pKa = 4.03ITSNGDD433 pKa = 3.0PTGITTRR440 pKa = 11.84GTATTTVITTVKK452 pKa = 6.89TTNGGSAVTYY462 pKa = 6.96TTTKK466 pKa = 10.1VVQPTQGSEE475 pKa = 3.85DD476 pKa = 4.35DD477 pKa = 4.16EE478 pKa = 5.35DD479 pKa = 4.88CEE481 pKa = 4.37EE482 pKa = 5.48YY483 pKa = 9.62YY484 pKa = 9.96TQTQTYY490 pKa = 6.87YY491 pKa = 7.96TTYY494 pKa = 7.66TTVINGATVTKK505 pKa = 10.51VVEE508 pKa = 4.12CDD510 pKa = 3.03EE511 pKa = 4.57CKK513 pKa = 10.35GTGSGSGSSGSGSSGSGSSGSGSYY537 pKa = 10.82GGGSSSGSGVDD548 pKa = 3.98EE549 pKa = 4.72DD550 pKa = 5.64CEE552 pKa = 4.68DD553 pKa = 4.62DD554 pKa = 4.19EE555 pKa = 5.6SSPSYY560 pKa = 11.14GSGSGSGTSYY570 pKa = 11.44GSGSGSSGSGSNYY583 pKa = 9.89GSGSGSSSGSGSGSGSTSKK602 pKa = 10.38PGYY605 pKa = 9.74LIPSDD610 pKa = 4.08DD611 pKa = 4.06QCDD614 pKa = 3.92EE615 pKa = 4.4DD616 pKa = 6.2CVVCDD621 pKa = 5.59DD622 pKa = 5.63SDD624 pKa = 6.34DD625 pKa = 4.87EE626 pKa = 4.96DD627 pKa = 5.29FDD629 pKa = 4.07WFGNDD634 pKa = 4.16TDD636 pKa = 4.9DD637 pKa = 5.39EE638 pKa = 5.37CDD640 pKa = 3.85ADD642 pKa = 5.54CDD644 pKa = 3.77EE645 pKa = 5.33CDD647 pKa = 3.68AGTSGSTSGSTSGSGSGSGSGSGSGSGSKK676 pKa = 10.14SGSGSGSGSGGSGSGSGSGGSGGAKK701 pKa = 9.26TYY703 pKa = 10.84VSTVRR708 pKa = 11.84TTAAGEE714 pKa = 4.37STWYY718 pKa = 8.1STSVSTSTHH727 pKa = 6.05SPDD730 pKa = 5.14DD731 pKa = 3.66EE732 pKa = 6.58CDD734 pKa = 3.76DD735 pKa = 4.42EE736 pKa = 5.62EE737 pKa = 5.18SATTTKK743 pKa = 10.73SATTTSGYY751 pKa = 11.39ANSTSKK757 pKa = 8.68QTGGFLADD765 pKa = 3.74LFGRR769 pKa = 11.84DD770 pKa = 3.72VEE772 pKa = 4.48ISSSAWANTSVTSAPVVEE790 pKa = 4.16QHH792 pKa = 6.8PSNGSRR798 pKa = 11.84QTSFSTALVALVVSIVAFTLII819 pKa = 4.01
Molecular weight: 78.48 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A5DQM9|A5DQM9_PICGU TAFII28 domain-containing protein OS=Meyerozyma guilliermondii (strain ATCC 6260 / CBS 566 / DSM 6381 / JCM 1539 / NBRC 10279 / NRRL Y-324) OX=294746 GN=PGUG_05580 PE=3 SV=2
MM1 pKa = 7.0YY2 pKa = 10.63VHH4 pKa = 7.36AKK6 pKa = 9.26TDD8 pKa = 3.27GEE10 pKa = 4.28MQKK13 pKa = 10.97KK14 pKa = 9.91KK15 pKa = 9.33MRR17 pKa = 11.84LWEE20 pKa = 3.69LHH22 pKa = 5.22GAINAGLTPILTSPYY37 pKa = 7.88GTKK40 pKa = 10.34ASEE43 pKa = 3.66ITRR46 pKa = 11.84FRR48 pKa = 11.84WCVNPPSNSILVGRR62 pKa = 11.84CMRR65 pKa = 11.84LICPDD70 pKa = 3.05HH71 pKa = 6.01SHH73 pKa = 6.72AVRR76 pKa = 11.84SCRR79 pKa = 11.84PSSTRR84 pKa = 11.84ARR86 pKa = 11.84ATTTARR92 pKa = 11.84DD93 pKa = 3.54AHH95 pKa = 6.57HH96 pKa = 6.64ATAGTRR102 pKa = 11.84RR103 pKa = 11.84WCPSILMDD111 pKa = 4.84LGVWVGKK118 pKa = 10.2RR119 pKa = 11.84LAGFVGVWACGVGPPTLLTNRR140 pKa = 11.84EE141 pKa = 4.03RR142 pKa = 11.84QVSIMTDD149 pKa = 3.2TAAYY153 pKa = 7.42EE154 pKa = 4.02YY155 pKa = 10.15PAIRR159 pKa = 11.84KK160 pKa = 8.49SFQRR164 pKa = 11.84FQIMFACRR172 pKa = 11.84ILIQHH177 pKa = 6.57RR178 pKa = 11.84RR179 pKa = 11.84AII181 pKa = 3.94
MM1 pKa = 7.0YY2 pKa = 10.63VHH4 pKa = 7.36AKK6 pKa = 9.26TDD8 pKa = 3.27GEE10 pKa = 4.28MQKK13 pKa = 10.97KK14 pKa = 9.91KK15 pKa = 9.33MRR17 pKa = 11.84LWEE20 pKa = 3.69LHH22 pKa = 5.22GAINAGLTPILTSPYY37 pKa = 7.88GTKK40 pKa = 10.34ASEE43 pKa = 3.66ITRR46 pKa = 11.84FRR48 pKa = 11.84WCVNPPSNSILVGRR62 pKa = 11.84CMRR65 pKa = 11.84LICPDD70 pKa = 3.05HH71 pKa = 6.01SHH73 pKa = 6.72AVRR76 pKa = 11.84SCRR79 pKa = 11.84PSSTRR84 pKa = 11.84ARR86 pKa = 11.84ATTTARR92 pKa = 11.84DD93 pKa = 3.54AHH95 pKa = 6.57HH96 pKa = 6.64ATAGTRR102 pKa = 11.84RR103 pKa = 11.84WCPSILMDD111 pKa = 4.84LGVWVGKK118 pKa = 10.2RR119 pKa = 11.84LAGFVGVWACGVGPPTLLTNRR140 pKa = 11.84EE141 pKa = 4.03RR142 pKa = 11.84QVSIMTDD149 pKa = 3.2TAAYY153 pKa = 7.42EE154 pKa = 4.02YY155 pKa = 10.15PAIRR159 pKa = 11.84KK160 pKa = 8.49SFQRR164 pKa = 11.84FQIMFACRR172 pKa = 11.84ILIQHH177 pKa = 6.57RR178 pKa = 11.84RR179 pKa = 11.84AII181 pKa = 3.94
Molecular weight: 20.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2761741 |
34 |
4897 |
466.6 |
52.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.352 ± 0.029 | 1.224 ± 0.011 |
5.751 ± 0.024 | 6.451 ± 0.029 |
4.421 ± 0.022 | 5.503 ± 0.035 |
2.318 ± 0.015 | 5.959 ± 0.021 |
6.508 ± 0.03 | 9.256 ± 0.034 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.098 ± 0.013 | 4.98 ± 0.019 |
4.705 ± 0.027 | 3.924 ± 0.022 |
4.717 ± 0.019 | 9.27 ± 0.044 |
5.713 ± 0.033 | 6.334 ± 0.026 |
1.114 ± 0.01 | 3.401 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
