
Salmonella phage FSLSP004
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; Peduovirinae; Stockinghallvirus; Salmonella virus FSLSP004
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
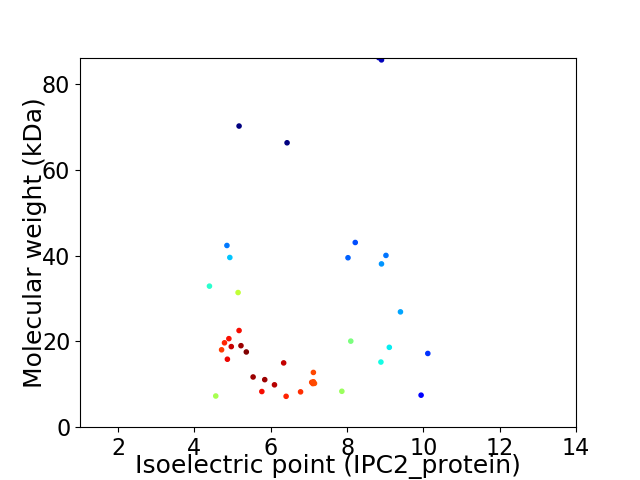
Virtual 2D-PAGE plot for 40 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
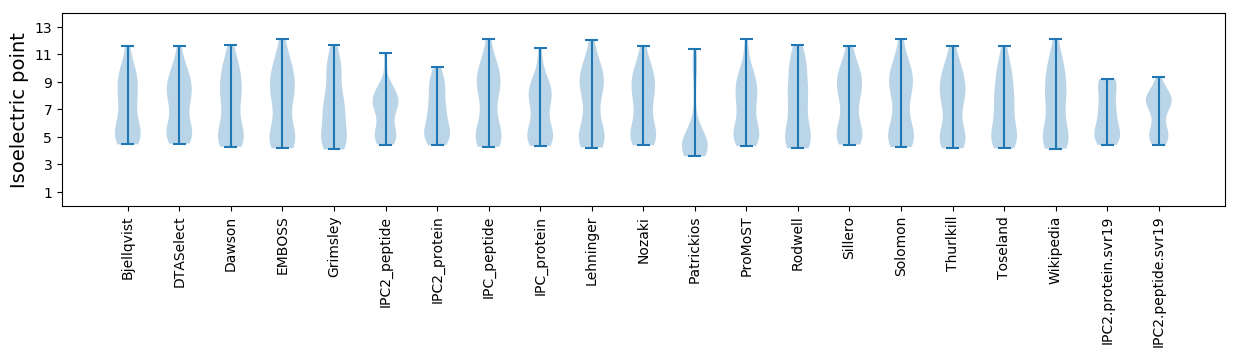
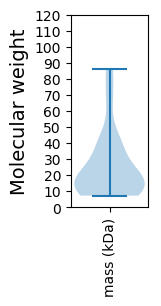
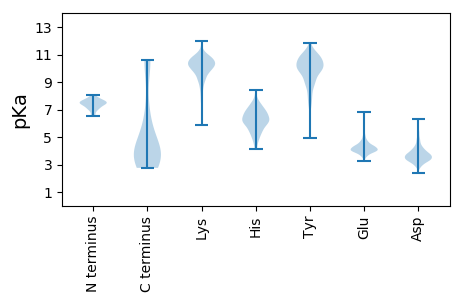
Protein with the lowest isoelectric point:
>tr|S4TNZ0|S4TNZ0_9CAUD Major tail tube protein OS=Salmonella phage FSLSP004 OX=1173769 GN=SP004_00105 PE=4 SV=1
MM1 pKa = 7.23ATVDD5 pKa = 4.28LSQLPVPDD13 pKa = 3.55VVEE16 pKa = 4.06EE17 pKa = 4.11LDD19 pKa = 3.72YY20 pKa = 10.57EE21 pKa = 4.61TILAEE26 pKa = 5.12RR27 pKa = 11.84IATLISLYY35 pKa = 10.45PEE37 pKa = 4.5DD38 pKa = 3.46QQEE41 pKa = 4.12AVARR45 pKa = 11.84TLALEE50 pKa = 4.08SEE52 pKa = 5.07PIVKK56 pKa = 10.22LLQEE60 pKa = 3.72NAYY63 pKa = 10.54RR64 pKa = 11.84EE65 pKa = 4.54VIWRR69 pKa = 11.84QRR71 pKa = 11.84VNEE74 pKa = 3.96AARR77 pKa = 11.84AVMLAYY83 pKa = 10.24AIDD86 pKa = 4.06SDD88 pKa = 4.56LDD90 pKa = 4.07NIGGNFSVEE99 pKa = 3.97RR100 pKa = 11.84LVVTPADD107 pKa = 3.87DD108 pKa = 3.72TTIPPTPAEE117 pKa = 4.27MEE119 pKa = 3.93LDD121 pKa = 2.96ADD123 pKa = 3.62YY124 pKa = 11.22RR125 pKa = 11.84LRR127 pKa = 11.84IQQAFEE133 pKa = 4.12GLSVAGPVGAYY144 pKa = 9.25QYY146 pKa = 10.44HH147 pKa = 6.16GRR149 pKa = 11.84SADD152 pKa = 3.24GRR154 pKa = 11.84VGDD157 pKa = 4.33ISVISPSPACVTISVLSRR175 pKa = 11.84EE176 pKa = 4.35NNGVASEE183 pKa = 4.1EE184 pKa = 3.82LLAIVRR190 pKa = 11.84NALNAEE196 pKa = 4.28DD197 pKa = 4.08VRR199 pKa = 11.84PVADD203 pKa = 3.2RR204 pKa = 11.84VTVQSAEE211 pKa = 3.77IVNYY215 pKa = 9.8QINATLYY222 pKa = 9.71LYY224 pKa = 8.88PGPEE228 pKa = 4.09SEE230 pKa = 5.15PIRR233 pKa = 11.84AAAEE237 pKa = 4.21AKK239 pKa = 10.05LKK241 pKa = 10.73AYY243 pKa = 9.88ISAQHH248 pKa = 6.43RR249 pKa = 11.84LGRR252 pKa = 11.84DD253 pKa = 2.91IRR255 pKa = 11.84KK256 pKa = 8.91SAIYY260 pKa = 9.91AALHH264 pKa = 5.3VEE266 pKa = 4.81GVQRR270 pKa = 11.84VEE272 pKa = 3.97LAAPVADD279 pKa = 2.95IVLDD283 pKa = 3.5NTQASFCTDD292 pKa = 3.12YY293 pKa = 11.51SLVIGGSDD301 pKa = 3.28EE302 pKa = 3.99
MM1 pKa = 7.23ATVDD5 pKa = 4.28LSQLPVPDD13 pKa = 3.55VVEE16 pKa = 4.06EE17 pKa = 4.11LDD19 pKa = 3.72YY20 pKa = 10.57EE21 pKa = 4.61TILAEE26 pKa = 5.12RR27 pKa = 11.84IATLISLYY35 pKa = 10.45PEE37 pKa = 4.5DD38 pKa = 3.46QQEE41 pKa = 4.12AVARR45 pKa = 11.84TLALEE50 pKa = 4.08SEE52 pKa = 5.07PIVKK56 pKa = 10.22LLQEE60 pKa = 3.72NAYY63 pKa = 10.54RR64 pKa = 11.84EE65 pKa = 4.54VIWRR69 pKa = 11.84QRR71 pKa = 11.84VNEE74 pKa = 3.96AARR77 pKa = 11.84AVMLAYY83 pKa = 10.24AIDD86 pKa = 4.06SDD88 pKa = 4.56LDD90 pKa = 4.07NIGGNFSVEE99 pKa = 3.97RR100 pKa = 11.84LVVTPADD107 pKa = 3.87DD108 pKa = 3.72TTIPPTPAEE117 pKa = 4.27MEE119 pKa = 3.93LDD121 pKa = 2.96ADD123 pKa = 3.62YY124 pKa = 11.22RR125 pKa = 11.84LRR127 pKa = 11.84IQQAFEE133 pKa = 4.12GLSVAGPVGAYY144 pKa = 9.25QYY146 pKa = 10.44HH147 pKa = 6.16GRR149 pKa = 11.84SADD152 pKa = 3.24GRR154 pKa = 11.84VGDD157 pKa = 4.33ISVISPSPACVTISVLSRR175 pKa = 11.84EE176 pKa = 4.35NNGVASEE183 pKa = 4.1EE184 pKa = 3.82LLAIVRR190 pKa = 11.84NALNAEE196 pKa = 4.28DD197 pKa = 4.08VRR199 pKa = 11.84PVADD203 pKa = 3.2RR204 pKa = 11.84VTVQSAEE211 pKa = 3.77IVNYY215 pKa = 9.8QINATLYY222 pKa = 9.71LYY224 pKa = 8.88PGPEE228 pKa = 4.09SEE230 pKa = 5.15PIRR233 pKa = 11.84AAAEE237 pKa = 4.21AKK239 pKa = 10.05LKK241 pKa = 10.73AYY243 pKa = 9.88ISAQHH248 pKa = 6.43RR249 pKa = 11.84LGRR252 pKa = 11.84DD253 pKa = 2.91IRR255 pKa = 11.84KK256 pKa = 8.91SAIYY260 pKa = 9.91AALHH264 pKa = 5.3VEE266 pKa = 4.81GVQRR270 pKa = 11.84VEE272 pKa = 3.97LAAPVADD279 pKa = 2.95IVLDD283 pKa = 3.5NTQASFCTDD292 pKa = 3.12YY293 pKa = 11.51SLVIGGSDD301 pKa = 3.28EE302 pKa = 3.99
Molecular weight: 32.86 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S4TP00|S4TP00_9CAUD DUF5405 domain-containing protein OS=Salmonella phage FSLSP004 OX=1173769 GN=SP004_00180 PE=4 SV=1
MM1 pKa = 7.53AFVDD5 pKa = 4.75GVVIMNEE12 pKa = 3.97PRR14 pKa = 11.84CIAQLLRR21 pKa = 11.84NEE23 pKa = 4.34SPRR26 pKa = 11.84AIDD29 pKa = 3.27FTITHH34 pKa = 6.18GKK36 pKa = 7.96GRR38 pKa = 11.84KK39 pKa = 9.25GIIIRR44 pKa = 11.84TKK46 pKa = 10.04KK47 pKa = 10.34QSPLKK52 pKa = 10.41KK53 pKa = 10.19ALTFLKK59 pKa = 10.34SRR61 pKa = 11.84RR62 pKa = 11.84VWKK65 pKa = 10.53
MM1 pKa = 7.53AFVDD5 pKa = 4.75GVVIMNEE12 pKa = 3.97PRR14 pKa = 11.84CIAQLLRR21 pKa = 11.84NEE23 pKa = 4.34SPRR26 pKa = 11.84AIDD29 pKa = 3.27FTITHH34 pKa = 6.18GKK36 pKa = 7.96GRR38 pKa = 11.84KK39 pKa = 9.25GIIIRR44 pKa = 11.84TKK46 pKa = 10.04KK47 pKa = 10.34QSPLKK52 pKa = 10.41KK53 pKa = 10.19ALTFLKK59 pKa = 10.34SRR61 pKa = 11.84RR62 pKa = 11.84VWKK65 pKa = 10.53
Molecular weight: 7.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
9148 |
63 |
813 |
228.7 |
25.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.319 ± 0.519 | 0.918 ± 0.151 |
5.805 ± 0.305 | 6.067 ± 0.29 |
3.268 ± 0.297 | 6.909 ± 0.458 |
1.957 ± 0.214 | 5.367 ± 0.274 |
5.619 ± 0.398 | 9.062 ± 0.279 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.613 ± 0.14 | 4.077 ± 0.259 |
4.154 ± 0.276 | 3.99 ± 0.254 |
6.439 ± 0.509 | 6.176 ± 0.335 |
6.362 ± 0.407 | 6.679 ± 0.388 |
1.443 ± 0.14 | 2.777 ± 0.172 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
