
Endozoicomonas elysicola
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Oceanospirillales; Endozoicomonadaceae; Endozoicomonas
Average proteome isoelectric point is 6.11
Get precalculated fractions of proteins
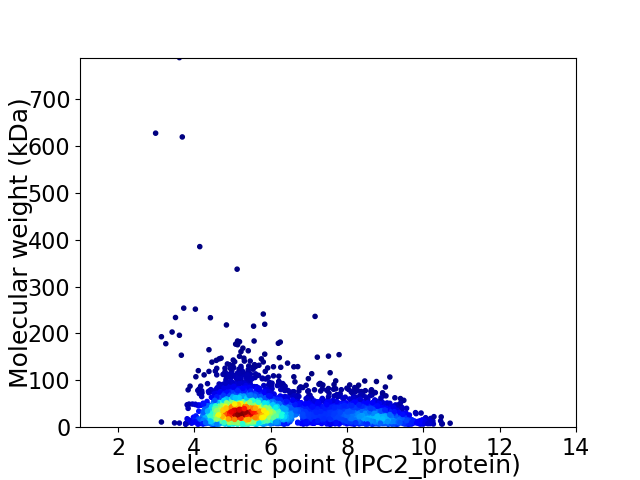
Virtual 2D-PAGE plot for 4147 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A081K8W0|A0A081K8W0_9GAMM Uncharacterized protein OS=Endozoicomonas elysicola OX=305900 GN=GV64_07395 PE=4 SV=1
MM1 pKa = 7.26NRR3 pKa = 11.84NDD5 pKa = 4.29LFRR8 pKa = 11.84KK9 pKa = 9.7SLLSTAVVAVLHH21 pKa = 6.14TYY23 pKa = 11.49SMGAAAACIPEE34 pKa = 4.35NVDD37 pKa = 3.26GVNTIIVDD45 pKa = 3.84DD46 pKa = 4.45GTDD49 pKa = 3.63CGDD52 pKa = 3.6ISITTPDD59 pKa = 3.71NVNHH63 pKa = 6.61IEE65 pKa = 4.01VEE67 pKa = 4.61GIVTGDD73 pKa = 3.03ITNATDD79 pKa = 4.25LSFLKK84 pKa = 10.38IEE86 pKa = 4.66AGDD89 pKa = 3.72VGEE92 pKa = 4.46GWIQGSVINQGDD104 pKa = 3.68ISHH107 pKa = 7.12HH108 pKa = 6.44DD109 pKa = 3.26VHH111 pKa = 6.59KK112 pKa = 11.04ASFSLSGPKK121 pKa = 9.87EE122 pKa = 4.07LLTVDD127 pKa = 4.44GDD129 pKa = 4.02VLNAGSATYY138 pKa = 8.84MQVEE142 pKa = 4.32HH143 pKa = 7.53ADD145 pKa = 3.16IGGDD149 pKa = 3.67IINSGQLTDD158 pKa = 5.54DD159 pKa = 4.08FLLVDD164 pKa = 3.72DD165 pKa = 5.08TIVYY169 pKa = 10.25GSVINTEE176 pKa = 3.97SGTIGSAEE184 pKa = 3.91NSVYY188 pKa = 10.81DD189 pKa = 5.63AILLNDD195 pKa = 3.74AVIKK199 pKa = 10.67EE200 pKa = 4.11GVINNGTITGEE211 pKa = 3.78NSGILISKK219 pKa = 9.98SEE221 pKa = 3.76ITGTQEE227 pKa = 3.58IEE229 pKa = 4.25GVLGAVVNNGDD240 pKa = 3.64INLGTGGKK248 pKa = 8.94EE249 pKa = 3.6YY250 pKa = 10.6AGRR253 pKa = 11.84GMFGIQITNAMPLDD267 pKa = 4.07APGGSEE273 pKa = 4.1DD274 pKa = 4.21QEE276 pKa = 4.16IATSIISGSVINTGNITVTDD296 pKa = 4.69GAPEE300 pKa = 4.12EE301 pKa = 4.56PQLQDD306 pKa = 3.43DD307 pKa = 4.47RR308 pKa = 11.84FGGIYY313 pKa = 9.66IGSEE317 pKa = 3.45WRR319 pKa = 11.84GKK321 pKa = 10.59EE322 pKa = 3.89DD323 pKa = 3.93DD324 pKa = 3.95SVAAHH329 pKa = 5.88VKK331 pKa = 10.6GNVINTGTIDD341 pKa = 3.65VSVTNTDD348 pKa = 2.96RR349 pKa = 11.84GEE351 pKa = 4.14TFGILIEE358 pKa = 4.05NAIVDD363 pKa = 4.09GTVGNIGIDD372 pKa = 3.62SLITTSRR379 pKa = 11.84EE380 pKa = 3.81GIIVVDD386 pKa = 4.58GSDD389 pKa = 3.02VGDD392 pKa = 3.67IVNEE396 pKa = 3.91GTITVTEE403 pKa = 4.63FWDD406 pKa = 4.82GIYY409 pKa = 10.46VDD411 pKa = 4.37SSSAASISNSGSITTTAGSGIYY433 pKa = 10.1LDD435 pKa = 3.77AAVVEE440 pKa = 4.55GTIRR444 pKa = 11.84NDD446 pKa = 3.62GQITANGAYY455 pKa = 9.22IEE457 pKa = 4.57YY458 pKa = 9.62PVGDD462 pKa = 4.1YY463 pKa = 10.98EE464 pKa = 4.47PASEE468 pKa = 4.27YY469 pKa = 11.06LDD471 pKa = 3.61GSGIFVSAYY480 pKa = 9.56GEE482 pKa = 4.14CDD484 pKa = 3.35EE485 pKa = 5.84EE486 pKa = 6.01YY487 pKa = 10.42DD488 pKa = 4.37DD489 pKa = 6.43CPITPSYY496 pKa = 10.0VGSIINGEE504 pKa = 3.99NGVINATEE512 pKa = 3.84AGIDD516 pKa = 3.43IDD518 pKa = 3.85HH519 pKa = 6.46SVIGSISNLGKK530 pKa = 10.12IVADD534 pKa = 3.54RR535 pKa = 11.84VGIYY539 pKa = 10.5AEE541 pKa = 4.16NGTIEE546 pKa = 4.55GNITNSGLIEE556 pKa = 4.62ADD558 pKa = 3.06GDD560 pKa = 4.49GISLMGVEE568 pKa = 4.26ATGIVNLGSITAHH581 pKa = 6.37NGLGIYY587 pKa = 10.44SSGSNLTDD595 pKa = 4.74GIYY598 pKa = 10.65NDD600 pKa = 4.3GQITSDD606 pKa = 3.67GPGIIAVSYY615 pKa = 10.46SPEE618 pKa = 3.71EE619 pKa = 3.92TVQVGFIANDD629 pKa = 3.33EE630 pKa = 4.43NGVITADD637 pKa = 3.5YY638 pKa = 10.23GPGIGAFAVIGEE650 pKa = 4.41SLANTGTINAEE661 pKa = 3.98SGIVAMLSTLTDD673 pKa = 3.56GVVNTGTINADD684 pKa = 2.8WVGIQLDD691 pKa = 3.94TTQAGTVANQGTINVSADD709 pKa = 3.01YY710 pKa = 10.54GAAIDD715 pKa = 3.85IRR717 pKa = 11.84GSKK720 pKa = 8.56LTTGVYY726 pKa = 11.01NNGTLNASEE735 pKa = 5.3GILASGYY742 pKa = 9.82PDD744 pKa = 4.48FGPVRR749 pKa = 11.84AAEE752 pKa = 4.62DD753 pKa = 4.3LPPPPPPPGPVPLEE767 pKa = 3.78EE768 pKa = 4.15TAPLSIGSVVNDD780 pKa = 3.33EE781 pKa = 4.97DD782 pKa = 4.39GVINATAEE790 pKa = 3.83DD791 pKa = 4.22SYY793 pKa = 11.75GIGLEE798 pKa = 3.99FVEE801 pKa = 5.55VVGLGNAGIINASYY815 pKa = 10.44GLNVYY820 pKa = 10.2EE821 pKa = 6.1SILTDD826 pKa = 4.59DD827 pKa = 4.02IVNTGTISASDD838 pKa = 3.19TGIAVEE844 pKa = 3.98NSQVNGNILNEE855 pKa = 3.9GSINGDD861 pKa = 3.2YY862 pKa = 10.69QGIHH866 pKa = 6.42INGKK870 pKa = 10.0LSGDD874 pKa = 3.45ILNTGTISSDD884 pKa = 2.77QGAAIYY890 pKa = 9.89TSFGVDD896 pKa = 2.51HH897 pKa = 7.46DD898 pKa = 4.82GSVEE902 pKa = 4.11NYY904 pKa = 10.43GSISGGNDD912 pKa = 3.17GEE914 pKa = 4.7TVTAIDD920 pKa = 3.92YY921 pKa = 10.6RR922 pKa = 11.84NAFAPLTLVQKK933 pKa = 10.7AGQIIGNIWGSSEE946 pKa = 4.47DD947 pKa = 3.97SGDD950 pKa = 3.27SAYY953 pKa = 9.79IDD955 pKa = 3.84GGNITGDD962 pKa = 3.27LHH964 pKa = 8.28DD965 pKa = 4.48IEE967 pKa = 6.1GIYY970 pKa = 10.36VDD972 pKa = 5.45GVTTLDD978 pKa = 3.52GNIYY982 pKa = 10.53GADD985 pKa = 4.52FINIEE990 pKa = 4.08SSGNLTVSGEE1000 pKa = 3.86ARR1002 pKa = 11.84SFEE1005 pKa = 4.6GYY1007 pKa = 10.78LNLYY1011 pKa = 9.88GGALSLNISNDD1022 pKa = 3.65TKK1024 pKa = 10.89PEE1026 pKa = 3.64EE1027 pKa = 4.4AMLSTEE1033 pKa = 3.93GEE1035 pKa = 4.33IYY1037 pKa = 10.62LSSDD1041 pKa = 2.55SRR1043 pKa = 11.84LIITASQNLEE1053 pKa = 4.19VNLDD1057 pKa = 3.73PVDD1060 pKa = 3.93YY1061 pKa = 10.64LALSGSALYY1070 pKa = 10.95NNGVTVEE1077 pKa = 4.27SGSAIITAEE1086 pKa = 3.93LAEE1089 pKa = 4.23VTEE1092 pKa = 5.19DD1093 pKa = 3.59SLSVTVATNDD1103 pKa = 3.12FGQVIEE1109 pKa = 4.57SVGATGNEE1117 pKa = 4.0VEE1119 pKa = 5.31VIDD1122 pKa = 6.23AYY1124 pKa = 8.54MQEE1127 pKa = 4.71YY1128 pKa = 9.22IASSGGEE1135 pKa = 4.09CNAICQAIEE1144 pKa = 3.77QAEE1147 pKa = 4.29TAEE1150 pKa = 4.08EE1151 pKa = 3.73LAAIAEE1157 pKa = 4.21EE1158 pKa = 4.2MVPDD1162 pKa = 3.66SSGVVGASQAAQSEE1176 pKa = 4.62TLGAVFKK1183 pKa = 10.61RR1184 pKa = 11.84IAGFRR1189 pKa = 11.84SSVSGVSSGDD1199 pKa = 3.47NIEE1202 pKa = 5.01PGSLWMQALASDD1214 pKa = 4.64GDD1216 pKa = 3.7QDD1218 pKa = 3.71AVQYY1222 pKa = 10.24QGNDD1226 pKa = 2.89FDD1228 pKa = 4.72GYY1230 pKa = 10.07SYY1232 pKa = 9.64RR1233 pKa = 11.84TRR1235 pKa = 11.84GFTIGADD1242 pKa = 3.39ADD1244 pKa = 4.05LSDD1247 pKa = 4.05TLTAGLAVTYY1257 pKa = 10.05GVTKK1261 pKa = 10.7SDD1263 pKa = 3.65VEE1265 pKa = 4.44GSANRR1270 pKa = 11.84SEE1272 pKa = 4.13TDD1274 pKa = 3.02SYY1276 pKa = 11.84LLTAYY1281 pKa = 10.0GSWRR1285 pKa = 11.84QQDD1288 pKa = 3.7YY1289 pKa = 11.3FMDD1292 pKa = 3.59SSIAFGVSRR1301 pKa = 11.84SDD1303 pKa = 3.25IEE1305 pKa = 4.36QYY1307 pKa = 8.73TANVKK1312 pKa = 8.7STADD1316 pKa = 3.07SDD1318 pKa = 4.08AAQVALRR1325 pKa = 11.84LVTGRR1330 pKa = 11.84TFAFNDD1336 pKa = 2.95NDD1338 pKa = 4.67TIVEE1342 pKa = 4.06PQVAFNYY1349 pKa = 10.41SRR1351 pKa = 11.84VDD1353 pKa = 3.42TNNYY1357 pKa = 7.54TLQPLNMEE1365 pKa = 4.43VLDD1368 pKa = 3.85QRR1370 pKa = 11.84IEE1372 pKa = 4.24TVEE1375 pKa = 3.91LGAGLRR1381 pKa = 11.84AMTAIDD1387 pKa = 3.78TKK1389 pKa = 10.68RR1390 pKa = 11.84GLLIPEE1396 pKa = 4.18VNLMAWHH1403 pKa = 7.35DD1404 pKa = 3.96FAADD1408 pKa = 3.82RR1409 pKa = 11.84GDD1411 pKa = 3.26TAARR1415 pKa = 11.84FLTGAGNSFVTTGAKK1430 pKa = 9.88PEE1432 pKa = 4.0QTNYY1436 pKa = 9.93QAGLGVQYY1444 pKa = 10.35WLNNNVSLSANYY1456 pKa = 9.44DD1457 pKa = 3.77RR1458 pKa = 11.84NWNSNFSADD1467 pKa = 2.9TWLANIRR1474 pKa = 11.84YY1475 pKa = 9.42DD1476 pKa = 3.56FF1477 pKa = 4.6
MM1 pKa = 7.26NRR3 pKa = 11.84NDD5 pKa = 4.29LFRR8 pKa = 11.84KK9 pKa = 9.7SLLSTAVVAVLHH21 pKa = 6.14TYY23 pKa = 11.49SMGAAAACIPEE34 pKa = 4.35NVDD37 pKa = 3.26GVNTIIVDD45 pKa = 3.84DD46 pKa = 4.45GTDD49 pKa = 3.63CGDD52 pKa = 3.6ISITTPDD59 pKa = 3.71NVNHH63 pKa = 6.61IEE65 pKa = 4.01VEE67 pKa = 4.61GIVTGDD73 pKa = 3.03ITNATDD79 pKa = 4.25LSFLKK84 pKa = 10.38IEE86 pKa = 4.66AGDD89 pKa = 3.72VGEE92 pKa = 4.46GWIQGSVINQGDD104 pKa = 3.68ISHH107 pKa = 7.12HH108 pKa = 6.44DD109 pKa = 3.26VHH111 pKa = 6.59KK112 pKa = 11.04ASFSLSGPKK121 pKa = 9.87EE122 pKa = 4.07LLTVDD127 pKa = 4.44GDD129 pKa = 4.02VLNAGSATYY138 pKa = 8.84MQVEE142 pKa = 4.32HH143 pKa = 7.53ADD145 pKa = 3.16IGGDD149 pKa = 3.67IINSGQLTDD158 pKa = 5.54DD159 pKa = 4.08FLLVDD164 pKa = 3.72DD165 pKa = 5.08TIVYY169 pKa = 10.25GSVINTEE176 pKa = 3.97SGTIGSAEE184 pKa = 3.91NSVYY188 pKa = 10.81DD189 pKa = 5.63AILLNDD195 pKa = 3.74AVIKK199 pKa = 10.67EE200 pKa = 4.11GVINNGTITGEE211 pKa = 3.78NSGILISKK219 pKa = 9.98SEE221 pKa = 3.76ITGTQEE227 pKa = 3.58IEE229 pKa = 4.25GVLGAVVNNGDD240 pKa = 3.64INLGTGGKK248 pKa = 8.94EE249 pKa = 3.6YY250 pKa = 10.6AGRR253 pKa = 11.84GMFGIQITNAMPLDD267 pKa = 4.07APGGSEE273 pKa = 4.1DD274 pKa = 4.21QEE276 pKa = 4.16IATSIISGSVINTGNITVTDD296 pKa = 4.69GAPEE300 pKa = 4.12EE301 pKa = 4.56PQLQDD306 pKa = 3.43DD307 pKa = 4.47RR308 pKa = 11.84FGGIYY313 pKa = 9.66IGSEE317 pKa = 3.45WRR319 pKa = 11.84GKK321 pKa = 10.59EE322 pKa = 3.89DD323 pKa = 3.93DD324 pKa = 3.95SVAAHH329 pKa = 5.88VKK331 pKa = 10.6GNVINTGTIDD341 pKa = 3.65VSVTNTDD348 pKa = 2.96RR349 pKa = 11.84GEE351 pKa = 4.14TFGILIEE358 pKa = 4.05NAIVDD363 pKa = 4.09GTVGNIGIDD372 pKa = 3.62SLITTSRR379 pKa = 11.84EE380 pKa = 3.81GIIVVDD386 pKa = 4.58GSDD389 pKa = 3.02VGDD392 pKa = 3.67IVNEE396 pKa = 3.91GTITVTEE403 pKa = 4.63FWDD406 pKa = 4.82GIYY409 pKa = 10.46VDD411 pKa = 4.37SSSAASISNSGSITTTAGSGIYY433 pKa = 10.1LDD435 pKa = 3.77AAVVEE440 pKa = 4.55GTIRR444 pKa = 11.84NDD446 pKa = 3.62GQITANGAYY455 pKa = 9.22IEE457 pKa = 4.57YY458 pKa = 9.62PVGDD462 pKa = 4.1YY463 pKa = 10.98EE464 pKa = 4.47PASEE468 pKa = 4.27YY469 pKa = 11.06LDD471 pKa = 3.61GSGIFVSAYY480 pKa = 9.56GEE482 pKa = 4.14CDD484 pKa = 3.35EE485 pKa = 5.84EE486 pKa = 6.01YY487 pKa = 10.42DD488 pKa = 4.37DD489 pKa = 6.43CPITPSYY496 pKa = 10.0VGSIINGEE504 pKa = 3.99NGVINATEE512 pKa = 3.84AGIDD516 pKa = 3.43IDD518 pKa = 3.85HH519 pKa = 6.46SVIGSISNLGKK530 pKa = 10.12IVADD534 pKa = 3.54RR535 pKa = 11.84VGIYY539 pKa = 10.5AEE541 pKa = 4.16NGTIEE546 pKa = 4.55GNITNSGLIEE556 pKa = 4.62ADD558 pKa = 3.06GDD560 pKa = 4.49GISLMGVEE568 pKa = 4.26ATGIVNLGSITAHH581 pKa = 6.37NGLGIYY587 pKa = 10.44SSGSNLTDD595 pKa = 4.74GIYY598 pKa = 10.65NDD600 pKa = 4.3GQITSDD606 pKa = 3.67GPGIIAVSYY615 pKa = 10.46SPEE618 pKa = 3.71EE619 pKa = 3.92TVQVGFIANDD629 pKa = 3.33EE630 pKa = 4.43NGVITADD637 pKa = 3.5YY638 pKa = 10.23GPGIGAFAVIGEE650 pKa = 4.41SLANTGTINAEE661 pKa = 3.98SGIVAMLSTLTDD673 pKa = 3.56GVVNTGTINADD684 pKa = 2.8WVGIQLDD691 pKa = 3.94TTQAGTVANQGTINVSADD709 pKa = 3.01YY710 pKa = 10.54GAAIDD715 pKa = 3.85IRR717 pKa = 11.84GSKK720 pKa = 8.56LTTGVYY726 pKa = 11.01NNGTLNASEE735 pKa = 5.3GILASGYY742 pKa = 9.82PDD744 pKa = 4.48FGPVRR749 pKa = 11.84AAEE752 pKa = 4.62DD753 pKa = 4.3LPPPPPPPGPVPLEE767 pKa = 3.78EE768 pKa = 4.15TAPLSIGSVVNDD780 pKa = 3.33EE781 pKa = 4.97DD782 pKa = 4.39GVINATAEE790 pKa = 3.83DD791 pKa = 4.22SYY793 pKa = 11.75GIGLEE798 pKa = 3.99FVEE801 pKa = 5.55VVGLGNAGIINASYY815 pKa = 10.44GLNVYY820 pKa = 10.2EE821 pKa = 6.1SILTDD826 pKa = 4.59DD827 pKa = 4.02IVNTGTISASDD838 pKa = 3.19TGIAVEE844 pKa = 3.98NSQVNGNILNEE855 pKa = 3.9GSINGDD861 pKa = 3.2YY862 pKa = 10.69QGIHH866 pKa = 6.42INGKK870 pKa = 10.0LSGDD874 pKa = 3.45ILNTGTISSDD884 pKa = 2.77QGAAIYY890 pKa = 9.89TSFGVDD896 pKa = 2.51HH897 pKa = 7.46DD898 pKa = 4.82GSVEE902 pKa = 4.11NYY904 pKa = 10.43GSISGGNDD912 pKa = 3.17GEE914 pKa = 4.7TVTAIDD920 pKa = 3.92YY921 pKa = 10.6RR922 pKa = 11.84NAFAPLTLVQKK933 pKa = 10.7AGQIIGNIWGSSEE946 pKa = 4.47DD947 pKa = 3.97SGDD950 pKa = 3.27SAYY953 pKa = 9.79IDD955 pKa = 3.84GGNITGDD962 pKa = 3.27LHH964 pKa = 8.28DD965 pKa = 4.48IEE967 pKa = 6.1GIYY970 pKa = 10.36VDD972 pKa = 5.45GVTTLDD978 pKa = 3.52GNIYY982 pKa = 10.53GADD985 pKa = 4.52FINIEE990 pKa = 4.08SSGNLTVSGEE1000 pKa = 3.86ARR1002 pKa = 11.84SFEE1005 pKa = 4.6GYY1007 pKa = 10.78LNLYY1011 pKa = 9.88GGALSLNISNDD1022 pKa = 3.65TKK1024 pKa = 10.89PEE1026 pKa = 3.64EE1027 pKa = 4.4AMLSTEE1033 pKa = 3.93GEE1035 pKa = 4.33IYY1037 pKa = 10.62LSSDD1041 pKa = 2.55SRR1043 pKa = 11.84LIITASQNLEE1053 pKa = 4.19VNLDD1057 pKa = 3.73PVDD1060 pKa = 3.93YY1061 pKa = 10.64LALSGSALYY1070 pKa = 10.95NNGVTVEE1077 pKa = 4.27SGSAIITAEE1086 pKa = 3.93LAEE1089 pKa = 4.23VTEE1092 pKa = 5.19DD1093 pKa = 3.59SLSVTVATNDD1103 pKa = 3.12FGQVIEE1109 pKa = 4.57SVGATGNEE1117 pKa = 4.0VEE1119 pKa = 5.31VIDD1122 pKa = 6.23AYY1124 pKa = 8.54MQEE1127 pKa = 4.71YY1128 pKa = 9.22IASSGGEE1135 pKa = 4.09CNAICQAIEE1144 pKa = 3.77QAEE1147 pKa = 4.29TAEE1150 pKa = 4.08EE1151 pKa = 3.73LAAIAEE1157 pKa = 4.21EE1158 pKa = 4.2MVPDD1162 pKa = 3.66SSGVVGASQAAQSEE1176 pKa = 4.62TLGAVFKK1183 pKa = 10.61RR1184 pKa = 11.84IAGFRR1189 pKa = 11.84SSVSGVSSGDD1199 pKa = 3.47NIEE1202 pKa = 5.01PGSLWMQALASDD1214 pKa = 4.64GDD1216 pKa = 3.7QDD1218 pKa = 3.71AVQYY1222 pKa = 10.24QGNDD1226 pKa = 2.89FDD1228 pKa = 4.72GYY1230 pKa = 10.07SYY1232 pKa = 9.64RR1233 pKa = 11.84TRR1235 pKa = 11.84GFTIGADD1242 pKa = 3.39ADD1244 pKa = 4.05LSDD1247 pKa = 4.05TLTAGLAVTYY1257 pKa = 10.05GVTKK1261 pKa = 10.7SDD1263 pKa = 3.65VEE1265 pKa = 4.44GSANRR1270 pKa = 11.84SEE1272 pKa = 4.13TDD1274 pKa = 3.02SYY1276 pKa = 11.84LLTAYY1281 pKa = 10.0GSWRR1285 pKa = 11.84QQDD1288 pKa = 3.7YY1289 pKa = 11.3FMDD1292 pKa = 3.59SSIAFGVSRR1301 pKa = 11.84SDD1303 pKa = 3.25IEE1305 pKa = 4.36QYY1307 pKa = 8.73TANVKK1312 pKa = 8.7STADD1316 pKa = 3.07SDD1318 pKa = 4.08AAQVALRR1325 pKa = 11.84LVTGRR1330 pKa = 11.84TFAFNDD1336 pKa = 2.95NDD1338 pKa = 4.67TIVEE1342 pKa = 4.06PQVAFNYY1349 pKa = 10.41SRR1351 pKa = 11.84VDD1353 pKa = 3.42TNNYY1357 pKa = 7.54TLQPLNMEE1365 pKa = 4.43VLDD1368 pKa = 3.85QRR1370 pKa = 11.84IEE1372 pKa = 4.24TVEE1375 pKa = 3.91LGAGLRR1381 pKa = 11.84AMTAIDD1387 pKa = 3.78TKK1389 pKa = 10.68RR1390 pKa = 11.84GLLIPEE1396 pKa = 4.18VNLMAWHH1403 pKa = 7.35DD1404 pKa = 3.96FAADD1408 pKa = 3.82RR1409 pKa = 11.84GDD1411 pKa = 3.26TAARR1415 pKa = 11.84FLTGAGNSFVTTGAKK1430 pKa = 9.88PEE1432 pKa = 4.0QTNYY1436 pKa = 9.93QAGLGVQYY1444 pKa = 10.35WLNNNVSLSANYY1456 pKa = 9.44DD1457 pKa = 3.77RR1458 pKa = 11.84NWNSNFSADD1467 pKa = 2.9TWLANIRR1474 pKa = 11.84YY1475 pKa = 9.42DD1476 pKa = 3.56FF1477 pKa = 4.6
Molecular weight: 153.59 kDa
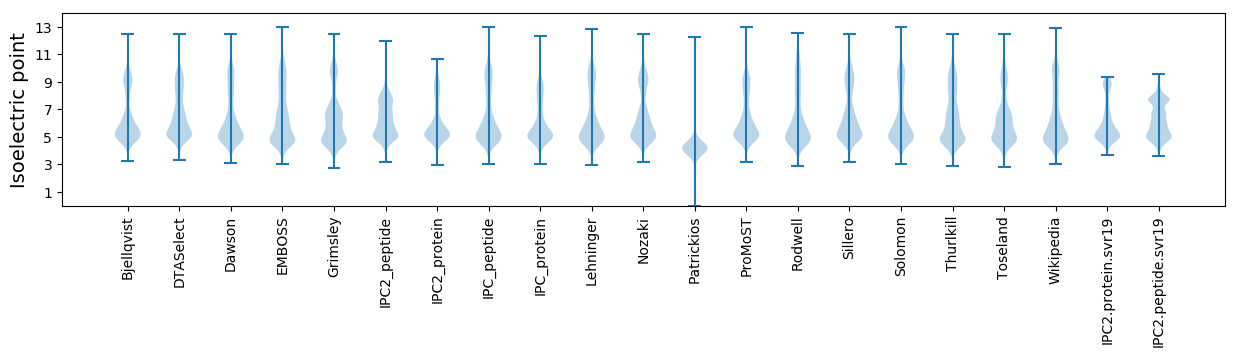
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A081K941|A0A081K941_9GAMM N-acetyltransferase domain-containing protein OS=Endozoicomonas elysicola OX=305900 GN=GV64_07870 PE=4 SV=1
MM1 pKa = 7.05VLSGMFLVIMLLHH14 pKa = 6.56LMVAMLIVFVSLFMGLMRR32 pKa = 11.84VFMPLWVATVLGVRR46 pKa = 11.84MAFFMLTIFQMITVVFIFNLRR67 pKa = 11.84VILGRR72 pKa = 11.84ALL74 pKa = 3.39
MM1 pKa = 7.05VLSGMFLVIMLLHH14 pKa = 6.56LMVAMLIVFVSLFMGLMRR32 pKa = 11.84VFMPLWVATVLGVRR46 pKa = 11.84MAFFMLTIFQMITVVFIFNLRR67 pKa = 11.84VILGRR72 pKa = 11.84ALL74 pKa = 3.39
Molecular weight: 8.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1412327 |
43 |
7482 |
340.6 |
37.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.196 ± 0.036 | 1.118 ± 0.017 |
5.66 ± 0.061 | 6.396 ± 0.037 |
3.972 ± 0.029 | 7.3 ± 0.072 |
2.272 ± 0.02 | 6.187 ± 0.029 |
4.979 ± 0.047 | 10.259 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.673 ± 0.024 | 4.124 ± 0.041 |
4.321 ± 0.036 | 4.316 ± 0.029 |
4.94 ± 0.041 | 7.016 ± 0.04 |
5.412 ± 0.059 | 6.682 ± 0.034 |
1.232 ± 0.015 | 2.944 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
