
Legionella londiniensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Legionellales; Legionellaceae; Legionella
Average proteome isoelectric point is 6.92
Get precalculated fractions of proteins
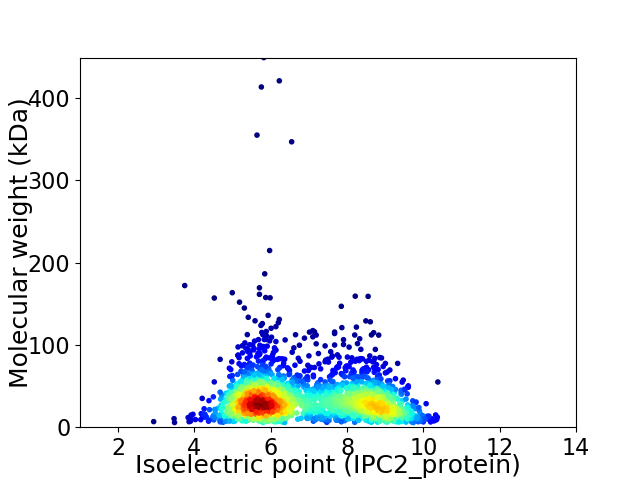
Virtual 2D-PAGE plot for 2172 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0W0VRV5|A0A0W0VRV5_9GAMM CTP synthase OS=Legionella londiniensis OX=45068 GN=pyrG PE=3 SV=1
MM1 pKa = 7.31AAIVSKK7 pKa = 10.92AVFFALPKK15 pKa = 9.27KK16 pKa = 8.69TATATPLNSQNIADD30 pKa = 5.54FIAGGYY36 pKa = 8.2WLFPSTLNDD45 pKa = 3.21QIYY48 pKa = 9.43YY49 pKa = 10.46QIFKK53 pKa = 10.97DD54 pKa = 4.6FALDD58 pKa = 3.58PNGRR62 pKa = 11.84YY63 pKa = 9.41SLEE66 pKa = 4.41LGDD69 pKa = 5.17GVILLSEE76 pKa = 4.25KK77 pKa = 11.03NIDD80 pKa = 3.77LSLILPPTAAGEE92 pKa = 4.1DD93 pKa = 3.73ANPILSYY100 pKa = 11.52VNGSEE105 pKa = 4.22VSSEE109 pKa = 3.93LVFIEE114 pKa = 5.35FINTLGQVEE123 pKa = 4.6VGFDD127 pKa = 3.22TRR129 pKa = 11.84ASFKK133 pKa = 10.6PFAYY137 pKa = 10.28DD138 pKa = 3.96LVPEE142 pKa = 4.29EE143 pKa = 4.23LTTYY147 pKa = 11.03ALRR150 pKa = 11.84FAFPDD155 pKa = 5.01GIISSPSLSVPTIPSAPLIHH175 pKa = 6.74LAADD179 pKa = 3.65TGISNTDD186 pKa = 4.42FITNNGSIIVSGLNPNATWQYY207 pKa = 10.99SVNNGPWITGNGNSFVLPEE226 pKa = 4.09GEE228 pKa = 4.29YY229 pKa = 10.8KK230 pKa = 10.63EE231 pKa = 4.67GAVKK235 pKa = 10.22VRR237 pKa = 11.84QINSEE242 pKa = 4.16GNISNTSSLKK252 pKa = 9.02PTVIDD257 pKa = 3.52TTHH260 pKa = 6.88PEE262 pKa = 3.4INIEE266 pKa = 4.17VNQTTEE272 pKa = 3.96DD273 pKa = 3.65NIVNIRR279 pKa = 11.84EE280 pKa = 4.09AGRR283 pKa = 11.84DD284 pKa = 3.18ITITGTVAGEE294 pKa = 3.99FNEE297 pKa = 4.28GDD299 pKa = 3.76TVTLTVNGKK308 pKa = 8.63EE309 pKa = 4.03FQGAVNAEE317 pKa = 4.18GLFSIAVPGRR327 pKa = 11.84DD328 pKa = 3.15LAADD332 pKa = 3.61ANLTIEE338 pKa = 4.45ASIATEE344 pKa = 3.68DD345 pKa = 3.56RR346 pKa = 11.84AGNKK350 pKa = 8.05ATASTDD356 pKa = 2.83HH357 pKa = 6.79TYY359 pKa = 10.86SINYY363 pKa = 9.26APEE366 pKa = 3.94TMEE369 pKa = 5.06DD370 pKa = 3.45LAHH373 pKa = 6.23VFEE376 pKa = 6.37GDD378 pKa = 3.78DD379 pKa = 4.19LIGSNVLSNDD389 pKa = 3.08TDD391 pKa = 3.81QNSSDD396 pKa = 3.55ILSVSLFSANSNGSGAEE413 pKa = 3.94TANGSNTITTLLGGTVTMNADD434 pKa = 3.01GTYY437 pKa = 9.5TYY439 pKa = 10.8SAPKK443 pKa = 9.96SVNNINGADD452 pKa = 3.74VVDD455 pKa = 3.78SFYY458 pKa = 11.63YY459 pKa = 10.15KK460 pKa = 10.82ASDD463 pKa = 4.35GYY465 pKa = 11.47NEE467 pKa = 4.32SDD469 pKa = 3.1WTKK472 pKa = 9.86VTINIQDD479 pKa = 3.65TSPTANPNEE488 pKa = 4.06ATVAYY493 pKa = 9.33QGSVSGNVTTNDD505 pKa = 4.03LFGQDD510 pKa = 3.32TAQLQSVTYY519 pKa = 10.26NHH521 pKa = 6.16ITYY524 pKa = 9.94DD525 pKa = 3.39QFVNGQLTINAANGVLTIHH544 pKa = 6.81QDD546 pKa = 2.94GTYY549 pKa = 9.3TYY551 pKa = 10.49QSNATLPRR559 pKa = 11.84AEE561 pKa = 4.08ITVGGSGGLSSWDD574 pKa = 3.03GVTLHH579 pKa = 6.49GFYY582 pKa = 10.67LGATEE587 pKa = 4.44PTPNSIGYY595 pKa = 8.7RR596 pKa = 11.84EE597 pKa = 4.38HH598 pKa = 6.47YY599 pKa = 10.01GLYY602 pKa = 9.56INSPGSVDD610 pKa = 3.22NSRR613 pKa = 11.84EE614 pKa = 3.6IDD616 pKa = 3.42SRR618 pKa = 11.84AVGDD622 pKa = 3.65TFKK625 pKa = 10.7TEE627 pKa = 4.23KK628 pKa = 10.32IVVEE632 pKa = 3.91FDD634 pKa = 3.73VNVTEE639 pKa = 4.38VTVGARR645 pKa = 11.84DD646 pKa = 3.85VQNHH650 pKa = 6.67DD651 pKa = 4.07SIHH654 pKa = 6.15WIAYY658 pKa = 9.39DD659 pKa = 3.64EE660 pKa = 4.83NGDD663 pKa = 3.81EE664 pKa = 4.25VATGIFTGEE673 pKa = 3.63NSAPGNDD680 pKa = 3.21HH681 pKa = 7.06TYY683 pKa = 10.27TFTITASEE691 pKa = 4.24AFRR694 pKa = 11.84SVAFMGHH701 pKa = 7.12DD702 pKa = 3.76GNDD705 pKa = 4.47DD706 pKa = 3.52FTLWNIKK713 pKa = 9.46YY714 pKa = 9.93IPAEE718 pKa = 4.22ANQDD722 pKa = 3.48VPPDD726 pKa = 3.29VFTYY730 pKa = 10.97VLVDD734 pKa = 3.62ADD736 pKa = 4.0GSTSQEE742 pKa = 3.75TTLTITHH749 pKa = 7.53DD750 pKa = 3.71STLGTTAVSDD760 pKa = 3.64EE761 pKa = 4.45SSVFEE766 pKa = 4.23SGLPQGTEE774 pKa = 3.53AGIASIVASGNLLANDD790 pKa = 3.81EE791 pKa = 4.48NVTPDD796 pKa = 3.2AAINEE801 pKa = 4.16ISFNGNTFAPVDD813 pKa = 3.81GVITIDD819 pKa = 3.56TPLGILTVYY828 pKa = 10.38TEE830 pKa = 4.89DD831 pKa = 5.07SVDD834 pKa = 3.3GHH836 pKa = 6.29NKK838 pKa = 9.74GDD840 pKa = 3.95YY841 pKa = 10.48EE842 pKa = 4.25YY843 pKa = 10.55TLQNASNEE851 pKa = 3.93GDD853 pKa = 3.59HH854 pKa = 6.93VSEE857 pKa = 4.35AFTYY861 pKa = 10.42QLINSSTGNASSSALTINIADD882 pKa = 4.29DD883 pKa = 3.77APRR886 pKa = 11.84GQDD889 pKa = 2.51IHH891 pKa = 8.24ASLNKK896 pKa = 9.82EE897 pKa = 3.91GSALTINMTLVIDD910 pKa = 4.11VSGSMNEE917 pKa = 3.73IDD919 pKa = 3.94SGEE922 pKa = 4.13TASRR926 pKa = 11.84LEE928 pKa = 3.93LAKK931 pKa = 10.59QSLEE935 pKa = 3.81EE936 pKa = 4.51LIRR939 pKa = 11.84TGDD942 pKa = 3.31EE943 pKa = 3.88AGTINIKK950 pKa = 10.33IIAFSTMAANSGWFVNNIEE969 pKa = 4.69AAIDD973 pKa = 3.81YY974 pKa = 10.83LNLLQANGYY983 pKa = 7.36TYY985 pKa = 11.16YY986 pKa = 11.19DD987 pKa = 3.32NALQEE992 pKa = 4.68VISSNGAPEE1001 pKa = 4.23PSADD1005 pKa = 3.2QHH1007 pKa = 6.42LLYY1010 pKa = 10.28FISDD1014 pKa = 4.19GEE1016 pKa = 4.25PTNNHH1021 pKa = 6.45SIANGNLTYY1030 pKa = 10.72NGYY1033 pKa = 10.47EE1034 pKa = 4.0DD1035 pKa = 3.83TEE1037 pKa = 4.02AWEE1040 pKa = 4.04QFITDD1045 pKa = 3.63SSIDD1049 pKa = 3.23KK1050 pKa = 10.75AFAVGFAGARR1060 pKa = 11.84SSALEE1065 pKa = 3.71PVAYY1069 pKa = 9.66PNGEE1073 pKa = 4.23DD1074 pKa = 3.34GGEE1077 pKa = 4.05VIIIDD1082 pKa = 4.18SASDD1086 pKa = 3.43LADD1089 pKa = 3.54ALINSLVDD1097 pKa = 3.3HH1098 pKa = 6.26VQTGTVINTEE1108 pKa = 3.95NASAEE1113 pKa = 4.1GFIIGADD1120 pKa = 3.53DD1121 pKa = 4.41AGSHH1125 pKa = 5.97IQSIMINHH1133 pKa = 5.59VVYY1136 pKa = 10.09EE1137 pKa = 4.34FNPAEE1142 pKa = 4.09PEE1144 pKa = 3.89IEE1146 pKa = 4.1VATGIKK1152 pKa = 10.31GATLTFNFLTGTYY1165 pKa = 10.02QYY1167 pKa = 10.44TVDD1170 pKa = 3.82SNEE1173 pKa = 4.04EE1174 pKa = 3.72IIAAEE1179 pKa = 4.2TFPITLVDD1187 pKa = 3.51GDD1189 pKa = 4.1GDD1191 pKa = 3.72EE1192 pKa = 4.4KK1193 pKa = 10.62TIHH1196 pKa = 6.26LTVQVNYY1203 pKa = 9.15EE1204 pKa = 3.82ASIDD1208 pKa = 3.84ANRR1211 pKa = 11.84DD1212 pKa = 3.69TIITNLTEE1220 pKa = 4.11GSLNFDD1226 pKa = 4.37ASALLNNDD1234 pKa = 3.19AGITNDD1240 pKa = 3.49VLISQVNNPKK1250 pKa = 10.68GGTVSLNNNMIQFNLNAQYY1269 pKa = 10.9SANDD1273 pKa = 3.58FNNTQYY1279 pKa = 11.88AKK1281 pKa = 10.17IVNEE1285 pKa = 4.19SEE1287 pKa = 4.18NNNTKK1292 pKa = 10.76LNADD1296 pKa = 4.03EE1297 pKa = 4.41ISRR1300 pKa = 11.84GSFKK1304 pKa = 11.27SNGLNNYY1311 pKa = 9.68SMTYY1315 pKa = 9.95RR1316 pKa = 11.84GAVSSNDD1323 pKa = 3.34ADD1325 pKa = 3.19WFMLSLAAGEE1335 pKa = 4.46LVHH1338 pKa = 7.67IDD1340 pKa = 4.47LNNTAYY1346 pKa = 10.95NIDD1349 pKa = 3.19IFAYY1353 pKa = 10.26DD1354 pKa = 3.75DD1355 pKa = 3.91NGNQLTQIFDD1365 pKa = 3.88YY1366 pKa = 10.48TFSSYY1371 pKa = 11.22YY1372 pKa = 8.26GTYY1375 pKa = 10.66GITNGLFAVQEE1386 pKa = 3.9EE1387 pKa = 4.77GNYY1390 pKa = 9.49YY1391 pKa = 10.25FQVAKK1396 pKa = 10.96NSGSSLNYY1404 pKa = 8.84EE1405 pKa = 4.08ANITIDD1411 pKa = 3.44TSNAQYY1417 pKa = 10.83QGFDD1421 pKa = 3.44YY1422 pKa = 10.74TVNDD1426 pKa = 3.83HH1427 pKa = 6.32NAVEE1431 pKa = 4.53ISAPVNIIVQEE1442 pKa = 4.18GNEE1445 pKa = 4.15LTGSEE1450 pKa = 4.21ASEE1453 pKa = 3.78ILIGSDD1459 pKa = 3.24GDD1461 pKa = 4.37DD1462 pKa = 4.3MINADD1467 pKa = 4.36GGDD1470 pKa = 3.66DD1471 pKa = 3.7VLFAGLGDD1479 pKa = 5.38DD1480 pKa = 4.26ILHH1483 pKa = 6.64GGDD1486 pKa = 4.33GNDD1489 pKa = 3.32TLFGGEE1495 pKa = 4.77GNDD1498 pKa = 3.47TMTGGDD1504 pKa = 3.77GQNTFVWKK1512 pKa = 10.68AGDD1515 pKa = 3.76DD1516 pKa = 3.66AGGSIDD1522 pKa = 4.34TITDD1526 pKa = 4.24FKK1528 pKa = 11.02LGQNGDD1534 pKa = 3.88VLDD1537 pKa = 5.03LSDD1540 pKa = 4.92LLVGEE1545 pKa = 4.78SNTAEE1550 pKa = 4.25SLDD1553 pKa = 3.59AYY1555 pKa = 11.39LNISYY1560 pKa = 10.89DD1561 pKa = 3.61NASNTSTVSVNPDD1574 pKa = 2.68GSASFSATQAIVLSGVDD1591 pKa = 3.59LTAGGTLQTDD1601 pKa = 3.55QAILDD1606 pKa = 3.75VLINNGNIVTDD1617 pKa = 4.25GGG1619 pKa = 3.73
MM1 pKa = 7.31AAIVSKK7 pKa = 10.92AVFFALPKK15 pKa = 9.27KK16 pKa = 8.69TATATPLNSQNIADD30 pKa = 5.54FIAGGYY36 pKa = 8.2WLFPSTLNDD45 pKa = 3.21QIYY48 pKa = 9.43YY49 pKa = 10.46QIFKK53 pKa = 10.97DD54 pKa = 4.6FALDD58 pKa = 3.58PNGRR62 pKa = 11.84YY63 pKa = 9.41SLEE66 pKa = 4.41LGDD69 pKa = 5.17GVILLSEE76 pKa = 4.25KK77 pKa = 11.03NIDD80 pKa = 3.77LSLILPPTAAGEE92 pKa = 4.1DD93 pKa = 3.73ANPILSYY100 pKa = 11.52VNGSEE105 pKa = 4.22VSSEE109 pKa = 3.93LVFIEE114 pKa = 5.35FINTLGQVEE123 pKa = 4.6VGFDD127 pKa = 3.22TRR129 pKa = 11.84ASFKK133 pKa = 10.6PFAYY137 pKa = 10.28DD138 pKa = 3.96LVPEE142 pKa = 4.29EE143 pKa = 4.23LTTYY147 pKa = 11.03ALRR150 pKa = 11.84FAFPDD155 pKa = 5.01GIISSPSLSVPTIPSAPLIHH175 pKa = 6.74LAADD179 pKa = 3.65TGISNTDD186 pKa = 4.42FITNNGSIIVSGLNPNATWQYY207 pKa = 10.99SVNNGPWITGNGNSFVLPEE226 pKa = 4.09GEE228 pKa = 4.29YY229 pKa = 10.8KK230 pKa = 10.63EE231 pKa = 4.67GAVKK235 pKa = 10.22VRR237 pKa = 11.84QINSEE242 pKa = 4.16GNISNTSSLKK252 pKa = 9.02PTVIDD257 pKa = 3.52TTHH260 pKa = 6.88PEE262 pKa = 3.4INIEE266 pKa = 4.17VNQTTEE272 pKa = 3.96DD273 pKa = 3.65NIVNIRR279 pKa = 11.84EE280 pKa = 4.09AGRR283 pKa = 11.84DD284 pKa = 3.18ITITGTVAGEE294 pKa = 3.99FNEE297 pKa = 4.28GDD299 pKa = 3.76TVTLTVNGKK308 pKa = 8.63EE309 pKa = 4.03FQGAVNAEE317 pKa = 4.18GLFSIAVPGRR327 pKa = 11.84DD328 pKa = 3.15LAADD332 pKa = 3.61ANLTIEE338 pKa = 4.45ASIATEE344 pKa = 3.68DD345 pKa = 3.56RR346 pKa = 11.84AGNKK350 pKa = 8.05ATASTDD356 pKa = 2.83HH357 pKa = 6.79TYY359 pKa = 10.86SINYY363 pKa = 9.26APEE366 pKa = 3.94TMEE369 pKa = 5.06DD370 pKa = 3.45LAHH373 pKa = 6.23VFEE376 pKa = 6.37GDD378 pKa = 3.78DD379 pKa = 4.19LIGSNVLSNDD389 pKa = 3.08TDD391 pKa = 3.81QNSSDD396 pKa = 3.55ILSVSLFSANSNGSGAEE413 pKa = 3.94TANGSNTITTLLGGTVTMNADD434 pKa = 3.01GTYY437 pKa = 9.5TYY439 pKa = 10.8SAPKK443 pKa = 9.96SVNNINGADD452 pKa = 3.74VVDD455 pKa = 3.78SFYY458 pKa = 11.63YY459 pKa = 10.15KK460 pKa = 10.82ASDD463 pKa = 4.35GYY465 pKa = 11.47NEE467 pKa = 4.32SDD469 pKa = 3.1WTKK472 pKa = 9.86VTINIQDD479 pKa = 3.65TSPTANPNEE488 pKa = 4.06ATVAYY493 pKa = 9.33QGSVSGNVTTNDD505 pKa = 4.03LFGQDD510 pKa = 3.32TAQLQSVTYY519 pKa = 10.26NHH521 pKa = 6.16ITYY524 pKa = 9.94DD525 pKa = 3.39QFVNGQLTINAANGVLTIHH544 pKa = 6.81QDD546 pKa = 2.94GTYY549 pKa = 9.3TYY551 pKa = 10.49QSNATLPRR559 pKa = 11.84AEE561 pKa = 4.08ITVGGSGGLSSWDD574 pKa = 3.03GVTLHH579 pKa = 6.49GFYY582 pKa = 10.67LGATEE587 pKa = 4.44PTPNSIGYY595 pKa = 8.7RR596 pKa = 11.84EE597 pKa = 4.38HH598 pKa = 6.47YY599 pKa = 10.01GLYY602 pKa = 9.56INSPGSVDD610 pKa = 3.22NSRR613 pKa = 11.84EE614 pKa = 3.6IDD616 pKa = 3.42SRR618 pKa = 11.84AVGDD622 pKa = 3.65TFKK625 pKa = 10.7TEE627 pKa = 4.23KK628 pKa = 10.32IVVEE632 pKa = 3.91FDD634 pKa = 3.73VNVTEE639 pKa = 4.38VTVGARR645 pKa = 11.84DD646 pKa = 3.85VQNHH650 pKa = 6.67DD651 pKa = 4.07SIHH654 pKa = 6.15WIAYY658 pKa = 9.39DD659 pKa = 3.64EE660 pKa = 4.83NGDD663 pKa = 3.81EE664 pKa = 4.25VATGIFTGEE673 pKa = 3.63NSAPGNDD680 pKa = 3.21HH681 pKa = 7.06TYY683 pKa = 10.27TFTITASEE691 pKa = 4.24AFRR694 pKa = 11.84SVAFMGHH701 pKa = 7.12DD702 pKa = 3.76GNDD705 pKa = 4.47DD706 pKa = 3.52FTLWNIKK713 pKa = 9.46YY714 pKa = 9.93IPAEE718 pKa = 4.22ANQDD722 pKa = 3.48VPPDD726 pKa = 3.29VFTYY730 pKa = 10.97VLVDD734 pKa = 3.62ADD736 pKa = 4.0GSTSQEE742 pKa = 3.75TTLTITHH749 pKa = 7.53DD750 pKa = 3.71STLGTTAVSDD760 pKa = 3.64EE761 pKa = 4.45SSVFEE766 pKa = 4.23SGLPQGTEE774 pKa = 3.53AGIASIVASGNLLANDD790 pKa = 3.81EE791 pKa = 4.48NVTPDD796 pKa = 3.2AAINEE801 pKa = 4.16ISFNGNTFAPVDD813 pKa = 3.81GVITIDD819 pKa = 3.56TPLGILTVYY828 pKa = 10.38TEE830 pKa = 4.89DD831 pKa = 5.07SVDD834 pKa = 3.3GHH836 pKa = 6.29NKK838 pKa = 9.74GDD840 pKa = 3.95YY841 pKa = 10.48EE842 pKa = 4.25YY843 pKa = 10.55TLQNASNEE851 pKa = 3.93GDD853 pKa = 3.59HH854 pKa = 6.93VSEE857 pKa = 4.35AFTYY861 pKa = 10.42QLINSSTGNASSSALTINIADD882 pKa = 4.29DD883 pKa = 3.77APRR886 pKa = 11.84GQDD889 pKa = 2.51IHH891 pKa = 8.24ASLNKK896 pKa = 9.82EE897 pKa = 3.91GSALTINMTLVIDD910 pKa = 4.11VSGSMNEE917 pKa = 3.73IDD919 pKa = 3.94SGEE922 pKa = 4.13TASRR926 pKa = 11.84LEE928 pKa = 3.93LAKK931 pKa = 10.59QSLEE935 pKa = 3.81EE936 pKa = 4.51LIRR939 pKa = 11.84TGDD942 pKa = 3.31EE943 pKa = 3.88AGTINIKK950 pKa = 10.33IIAFSTMAANSGWFVNNIEE969 pKa = 4.69AAIDD973 pKa = 3.81YY974 pKa = 10.83LNLLQANGYY983 pKa = 7.36TYY985 pKa = 11.16YY986 pKa = 11.19DD987 pKa = 3.32NALQEE992 pKa = 4.68VISSNGAPEE1001 pKa = 4.23PSADD1005 pKa = 3.2QHH1007 pKa = 6.42LLYY1010 pKa = 10.28FISDD1014 pKa = 4.19GEE1016 pKa = 4.25PTNNHH1021 pKa = 6.45SIANGNLTYY1030 pKa = 10.72NGYY1033 pKa = 10.47EE1034 pKa = 4.0DD1035 pKa = 3.83TEE1037 pKa = 4.02AWEE1040 pKa = 4.04QFITDD1045 pKa = 3.63SSIDD1049 pKa = 3.23KK1050 pKa = 10.75AFAVGFAGARR1060 pKa = 11.84SSALEE1065 pKa = 3.71PVAYY1069 pKa = 9.66PNGEE1073 pKa = 4.23DD1074 pKa = 3.34GGEE1077 pKa = 4.05VIIIDD1082 pKa = 4.18SASDD1086 pKa = 3.43LADD1089 pKa = 3.54ALINSLVDD1097 pKa = 3.3HH1098 pKa = 6.26VQTGTVINTEE1108 pKa = 3.95NASAEE1113 pKa = 4.1GFIIGADD1120 pKa = 3.53DD1121 pKa = 4.41AGSHH1125 pKa = 5.97IQSIMINHH1133 pKa = 5.59VVYY1136 pKa = 10.09EE1137 pKa = 4.34FNPAEE1142 pKa = 4.09PEE1144 pKa = 3.89IEE1146 pKa = 4.1VATGIKK1152 pKa = 10.31GATLTFNFLTGTYY1165 pKa = 10.02QYY1167 pKa = 10.44TVDD1170 pKa = 3.82SNEE1173 pKa = 4.04EE1174 pKa = 3.72IIAAEE1179 pKa = 4.2TFPITLVDD1187 pKa = 3.51GDD1189 pKa = 4.1GDD1191 pKa = 3.72EE1192 pKa = 4.4KK1193 pKa = 10.62TIHH1196 pKa = 6.26LTVQVNYY1203 pKa = 9.15EE1204 pKa = 3.82ASIDD1208 pKa = 3.84ANRR1211 pKa = 11.84DD1212 pKa = 3.69TIITNLTEE1220 pKa = 4.11GSLNFDD1226 pKa = 4.37ASALLNNDD1234 pKa = 3.19AGITNDD1240 pKa = 3.49VLISQVNNPKK1250 pKa = 10.68GGTVSLNNNMIQFNLNAQYY1269 pKa = 10.9SANDD1273 pKa = 3.58FNNTQYY1279 pKa = 11.88AKK1281 pKa = 10.17IVNEE1285 pKa = 4.19SEE1287 pKa = 4.18NNNTKK1292 pKa = 10.76LNADD1296 pKa = 4.03EE1297 pKa = 4.41ISRR1300 pKa = 11.84GSFKK1304 pKa = 11.27SNGLNNYY1311 pKa = 9.68SMTYY1315 pKa = 9.95RR1316 pKa = 11.84GAVSSNDD1323 pKa = 3.34ADD1325 pKa = 3.19WFMLSLAAGEE1335 pKa = 4.46LVHH1338 pKa = 7.67IDD1340 pKa = 4.47LNNTAYY1346 pKa = 10.95NIDD1349 pKa = 3.19IFAYY1353 pKa = 10.26DD1354 pKa = 3.75DD1355 pKa = 3.91NGNQLTQIFDD1365 pKa = 3.88YY1366 pKa = 10.48TFSSYY1371 pKa = 11.22YY1372 pKa = 8.26GTYY1375 pKa = 10.66GITNGLFAVQEE1386 pKa = 3.9EE1387 pKa = 4.77GNYY1390 pKa = 9.49YY1391 pKa = 10.25FQVAKK1396 pKa = 10.96NSGSSLNYY1404 pKa = 8.84EE1405 pKa = 4.08ANITIDD1411 pKa = 3.44TSNAQYY1417 pKa = 10.83QGFDD1421 pKa = 3.44YY1422 pKa = 10.74TVNDD1426 pKa = 3.83HH1427 pKa = 6.32NAVEE1431 pKa = 4.53ISAPVNIIVQEE1442 pKa = 4.18GNEE1445 pKa = 4.15LTGSEE1450 pKa = 4.21ASEE1453 pKa = 3.78ILIGSDD1459 pKa = 3.24GDD1461 pKa = 4.37DD1462 pKa = 4.3MINADD1467 pKa = 4.36GGDD1470 pKa = 3.66DD1471 pKa = 3.7VLFAGLGDD1479 pKa = 5.38DD1480 pKa = 4.26ILHH1483 pKa = 6.64GGDD1486 pKa = 4.33GNDD1489 pKa = 3.32TLFGGEE1495 pKa = 4.77GNDD1498 pKa = 3.47TMTGGDD1504 pKa = 3.77GQNTFVWKK1512 pKa = 10.68AGDD1515 pKa = 3.76DD1516 pKa = 3.66AGGSIDD1522 pKa = 4.34TITDD1526 pKa = 4.24FKK1528 pKa = 11.02LGQNGDD1534 pKa = 3.88VLDD1537 pKa = 5.03LSDD1540 pKa = 4.92LLVGEE1545 pKa = 4.78SNTAEE1550 pKa = 4.25SLDD1553 pKa = 3.59AYY1555 pKa = 11.39LNISYY1560 pKa = 10.89DD1561 pKa = 3.61NASNTSTVSVNPDD1574 pKa = 2.68GSASFSATQAIVLSGVDD1591 pKa = 3.59LTAGGTLQTDD1601 pKa = 3.55QAILDD1606 pKa = 3.75VLINNGNIVTDD1617 pKa = 4.25GGG1619 pKa = 3.73
Molecular weight: 172.22 kDa
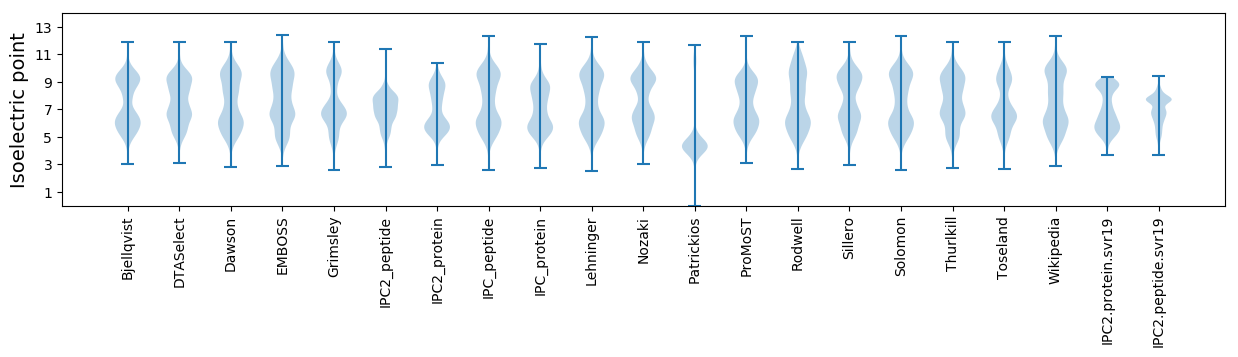
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0W0VT33|A0A0W0VT33_9GAMM 3-hydroxymyristoyl-ACP dehydratase OS=Legionella londiniensis OX=45068 GN=fabA PE=4 SV=1
MM1 pKa = 7.55ALIKK5 pKa = 10.53SKK7 pKa = 9.07PTSPGKK13 pKa = 10.09RR14 pKa = 11.84GEE16 pKa = 3.61IRR18 pKa = 11.84VRR20 pKa = 11.84HH21 pKa = 5.9DD22 pKa = 3.38VYY24 pKa = 11.16KK25 pKa = 10.86GDD27 pKa = 4.54PFPGLISTVKK37 pKa = 9.51KK38 pKa = 9.15TGGRR42 pKa = 11.84NNQGRR47 pKa = 11.84ITVRR51 pKa = 11.84HH52 pKa = 5.83IGGGTRR58 pKa = 11.84VQYY61 pKa = 10.79RR62 pKa = 11.84HH63 pKa = 6.6IDD65 pKa = 3.68FKK67 pKa = 11.19RR68 pKa = 11.84NKK70 pKa = 9.81DD71 pKa = 3.49GIEE74 pKa = 3.79ARR76 pKa = 11.84VEE78 pKa = 4.11RR79 pKa = 11.84IEE81 pKa = 3.97YY82 pKa = 10.37DD83 pKa = 3.49PNRR86 pKa = 11.84SALIALIVYY95 pKa = 10.06KK96 pKa = 10.6DD97 pKa = 3.22GEE99 pKa = 4.23RR100 pKa = 11.84RR101 pKa = 11.84YY102 pKa = 10.08IIAPAGLALNDD113 pKa = 4.01TVVSGDD119 pKa = 4.09DD120 pKa = 3.7APISTGNALPLKK132 pKa = 8.99NIPVGTTVHH141 pKa = 6.24CVEE144 pKa = 5.21LKK146 pKa = 10.19PGCGAQLLRR155 pKa = 11.84SAGCSGQIVAKK166 pKa = 9.68EE167 pKa = 3.95GAYY170 pKa = 8.78STLKK174 pKa = 10.45LRR176 pKa = 11.84SGEE179 pKa = 3.95MRR181 pKa = 11.84KK182 pKa = 9.5VLSACRR188 pKa = 11.84ATVGEE193 pKa = 4.46VSNSEE198 pKa = 4.06HH199 pKa = 6.74NLRR202 pKa = 11.84ALGKK206 pKa = 10.04AGASRR211 pKa = 11.84WRR213 pKa = 11.84GKK215 pKa = 10.27RR216 pKa = 11.84PTVRR220 pKa = 11.84GVAMNPVDD228 pKa = 3.85HH229 pKa = 6.82PHH231 pKa = 6.77GGGEE235 pKa = 4.38GKK237 pKa = 9.02TSGGRR242 pKa = 11.84HH243 pKa = 5.83PVTPWGIPTKK253 pKa = 10.33GYY255 pKa = 7.47KK256 pKa = 7.77TRR258 pKa = 11.84RR259 pKa = 11.84NKK261 pKa = 9.32RR262 pKa = 11.84TSKK265 pKa = 10.29YY266 pKa = 9.53IVRR269 pKa = 11.84GRR271 pKa = 11.84KK272 pKa = 8.88NKK274 pKa = 10.12
MM1 pKa = 7.55ALIKK5 pKa = 10.53SKK7 pKa = 9.07PTSPGKK13 pKa = 10.09RR14 pKa = 11.84GEE16 pKa = 3.61IRR18 pKa = 11.84VRR20 pKa = 11.84HH21 pKa = 5.9DD22 pKa = 3.38VYY24 pKa = 11.16KK25 pKa = 10.86GDD27 pKa = 4.54PFPGLISTVKK37 pKa = 9.51KK38 pKa = 9.15TGGRR42 pKa = 11.84NNQGRR47 pKa = 11.84ITVRR51 pKa = 11.84HH52 pKa = 5.83IGGGTRR58 pKa = 11.84VQYY61 pKa = 10.79RR62 pKa = 11.84HH63 pKa = 6.6IDD65 pKa = 3.68FKK67 pKa = 11.19RR68 pKa = 11.84NKK70 pKa = 9.81DD71 pKa = 3.49GIEE74 pKa = 3.79ARR76 pKa = 11.84VEE78 pKa = 4.11RR79 pKa = 11.84IEE81 pKa = 3.97YY82 pKa = 10.37DD83 pKa = 3.49PNRR86 pKa = 11.84SALIALIVYY95 pKa = 10.06KK96 pKa = 10.6DD97 pKa = 3.22GEE99 pKa = 4.23RR100 pKa = 11.84RR101 pKa = 11.84YY102 pKa = 10.08IIAPAGLALNDD113 pKa = 4.01TVVSGDD119 pKa = 4.09DD120 pKa = 3.7APISTGNALPLKK132 pKa = 8.99NIPVGTTVHH141 pKa = 6.24CVEE144 pKa = 5.21LKK146 pKa = 10.19PGCGAQLLRR155 pKa = 11.84SAGCSGQIVAKK166 pKa = 9.68EE167 pKa = 3.95GAYY170 pKa = 8.78STLKK174 pKa = 10.45LRR176 pKa = 11.84SGEE179 pKa = 3.95MRR181 pKa = 11.84KK182 pKa = 9.5VLSACRR188 pKa = 11.84ATVGEE193 pKa = 4.46VSNSEE198 pKa = 4.06HH199 pKa = 6.74NLRR202 pKa = 11.84ALGKK206 pKa = 10.04AGASRR211 pKa = 11.84WRR213 pKa = 11.84GKK215 pKa = 10.27RR216 pKa = 11.84PTVRR220 pKa = 11.84GVAMNPVDD228 pKa = 3.85HH229 pKa = 6.82PHH231 pKa = 6.77GGGEE235 pKa = 4.38GKK237 pKa = 9.02TSGGRR242 pKa = 11.84HH243 pKa = 5.83PVTPWGIPTKK253 pKa = 10.33GYY255 pKa = 7.47KK256 pKa = 7.77TRR258 pKa = 11.84RR259 pKa = 11.84NKK261 pKa = 9.32RR262 pKa = 11.84TSKK265 pKa = 10.29YY266 pKa = 9.53IVRR269 pKa = 11.84GRR271 pKa = 11.84KK272 pKa = 8.88NKK274 pKa = 10.12
Molecular weight: 29.71 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
722513 |
51 |
3879 |
332.6 |
37.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.372 ± 0.056 | 1.188 ± 0.018 |
4.7 ± 0.04 | 6.336 ± 0.054 |
4.471 ± 0.038 | 6.208 ± 0.044 |
2.595 ± 0.026 | 7.254 ± 0.047 |
6.19 ± 0.058 | 11.267 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.396 ± 0.021 | 4.372 ± 0.041 |
4.137 ± 0.034 | 4.593 ± 0.04 |
4.794 ± 0.039 | 6.081 ± 0.031 |
4.715 ± 0.033 | 5.787 ± 0.048 |
1.143 ± 0.02 | 3.393 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
