
Hyunsoonleella jejuensis
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Hyunsoonleella
Average proteome isoelectric point is 6.56
Get precalculated fractions of proteins
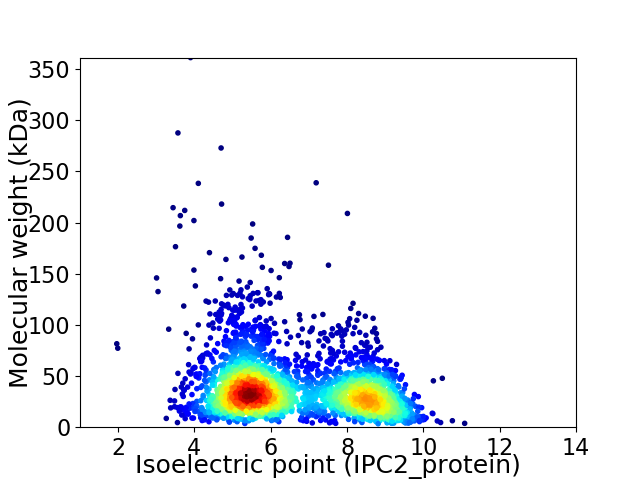
Virtual 2D-PAGE plot for 2939 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H9B3U9|A0A1H9B3U9_9FLAO Exo-beta-1 3-glucanase GH17 family OS=Hyunsoonleella jejuensis OX=419940 GN=SAMN05421824_0444 PE=4 SV=1
MM1 pKa = 7.08MKK3 pKa = 10.45KK4 pKa = 8.36ITLLLLTFLSFSLGFSQTEE23 pKa = 3.99LLNLSFDD30 pKa = 3.92AAGSEE35 pKa = 4.95SVWTPIADD43 pKa = 3.63AASNPSEE50 pKa = 4.06VNIAFDD56 pKa = 4.51ANGNPTGATLLSGINTSDD74 pKa = 3.28VAGRR78 pKa = 11.84AYY80 pKa = 9.75IFRR83 pKa = 11.84YY84 pKa = 10.17NNASFDD90 pKa = 3.81FGTSGSVSISMDD102 pKa = 3.32VKK104 pKa = 10.49IDD106 pKa = 3.45AALTGTNLVFEE117 pKa = 4.58TQVSKK122 pKa = 11.35VGGGVVVVTHH132 pKa = 6.56NNVQNDD138 pKa = 3.68VAVGGGWIPLTFDD151 pKa = 3.19MTPNPAEE158 pKa = 4.04FNNAGTEE165 pKa = 4.17LYY167 pKa = 10.45FYY169 pKa = 11.23FNMAAGAFVGAGGTFLVDD187 pKa = 3.79NIVVTGGAPAEE198 pKa = 4.17PTCDD202 pKa = 3.58DD203 pKa = 3.44GLMNGDD209 pKa = 3.92EE210 pKa = 4.38TGVDD214 pKa = 4.11CGGPDD219 pKa = 3.58CSACIEE225 pKa = 4.91DD226 pKa = 4.17PTDD229 pKa = 3.83APSEE233 pKa = 4.01LASTGTDD240 pKa = 3.26LYY242 pKa = 11.05AYY244 pKa = 9.88SGLSAVGEE252 pKa = 4.07SDD254 pKa = 4.25LPGFNFTAFNGGVTISEE271 pKa = 4.3EE272 pKa = 4.16TLSGNKK278 pKa = 9.17VGKK281 pKa = 9.4LANLDD286 pKa = 3.83FFGSGWTPIDD296 pKa = 3.2VTTAYY301 pKa = 9.64TYY303 pKa = 11.38VHH305 pKa = 7.04LDD307 pKa = 3.34YY308 pKa = 11.13YY309 pKa = 10.94AVSGTWFEE317 pKa = 4.67FFLIDD322 pKa = 4.73DD323 pKa = 4.38SLSATICCGSGEE335 pKa = 4.1EE336 pKa = 3.88PRR338 pKa = 11.84YY339 pKa = 10.28RR340 pKa = 11.84FGPAPSGQDD349 pKa = 3.13EE350 pKa = 4.41PLVLGAWTSVFIPLSDD366 pKa = 3.94FANYY370 pKa = 9.63AALTNGTWDD379 pKa = 3.41GTDD382 pKa = 2.81IKK384 pKa = 9.51EE385 pKa = 4.57TKK387 pKa = 8.89FTSDD391 pKa = 2.4GGTIYY396 pKa = 10.65FDD398 pKa = 3.97NIYY401 pKa = 10.49FSTSNVLGVDD411 pKa = 3.83DD412 pKa = 5.26FEE414 pKa = 4.61STSFSVYY421 pKa = 10.28PNPTQEE427 pKa = 3.43SWTIKK432 pKa = 10.09AQNTKK437 pKa = 8.11ITNIKK442 pKa = 10.18VYY444 pKa = 10.67DD445 pKa = 3.78VLGKK449 pKa = 10.14NVMMLSPNTIEE460 pKa = 4.17TKK462 pKa = 10.11IDD464 pKa = 3.31GASLMKK470 pKa = 10.58GIYY473 pKa = 8.37FAKK476 pKa = 10.07IEE478 pKa = 4.47TVSGTSSVKK487 pKa = 10.21LVKK490 pKa = 10.26QQ491 pKa = 3.39
MM1 pKa = 7.08MKK3 pKa = 10.45KK4 pKa = 8.36ITLLLLTFLSFSLGFSQTEE23 pKa = 3.99LLNLSFDD30 pKa = 3.92AAGSEE35 pKa = 4.95SVWTPIADD43 pKa = 3.63AASNPSEE50 pKa = 4.06VNIAFDD56 pKa = 4.51ANGNPTGATLLSGINTSDD74 pKa = 3.28VAGRR78 pKa = 11.84AYY80 pKa = 9.75IFRR83 pKa = 11.84YY84 pKa = 10.17NNASFDD90 pKa = 3.81FGTSGSVSISMDD102 pKa = 3.32VKK104 pKa = 10.49IDD106 pKa = 3.45AALTGTNLVFEE117 pKa = 4.58TQVSKK122 pKa = 11.35VGGGVVVVTHH132 pKa = 6.56NNVQNDD138 pKa = 3.68VAVGGGWIPLTFDD151 pKa = 3.19MTPNPAEE158 pKa = 4.04FNNAGTEE165 pKa = 4.17LYY167 pKa = 10.45FYY169 pKa = 11.23FNMAAGAFVGAGGTFLVDD187 pKa = 3.79NIVVTGGAPAEE198 pKa = 4.17PTCDD202 pKa = 3.58DD203 pKa = 3.44GLMNGDD209 pKa = 3.92EE210 pKa = 4.38TGVDD214 pKa = 4.11CGGPDD219 pKa = 3.58CSACIEE225 pKa = 4.91DD226 pKa = 4.17PTDD229 pKa = 3.83APSEE233 pKa = 4.01LASTGTDD240 pKa = 3.26LYY242 pKa = 11.05AYY244 pKa = 9.88SGLSAVGEE252 pKa = 4.07SDD254 pKa = 4.25LPGFNFTAFNGGVTISEE271 pKa = 4.3EE272 pKa = 4.16TLSGNKK278 pKa = 9.17VGKK281 pKa = 9.4LANLDD286 pKa = 3.83FFGSGWTPIDD296 pKa = 3.2VTTAYY301 pKa = 9.64TYY303 pKa = 11.38VHH305 pKa = 7.04LDD307 pKa = 3.34YY308 pKa = 11.13YY309 pKa = 10.94AVSGTWFEE317 pKa = 4.67FFLIDD322 pKa = 4.73DD323 pKa = 4.38SLSATICCGSGEE335 pKa = 4.1EE336 pKa = 3.88PRR338 pKa = 11.84YY339 pKa = 10.28RR340 pKa = 11.84FGPAPSGQDD349 pKa = 3.13EE350 pKa = 4.41PLVLGAWTSVFIPLSDD366 pKa = 3.94FANYY370 pKa = 9.63AALTNGTWDD379 pKa = 3.41GTDD382 pKa = 2.81IKK384 pKa = 9.51EE385 pKa = 4.57TKK387 pKa = 8.89FTSDD391 pKa = 2.4GGTIYY396 pKa = 10.65FDD398 pKa = 3.97NIYY401 pKa = 10.49FSTSNVLGVDD411 pKa = 3.83DD412 pKa = 5.26FEE414 pKa = 4.61STSFSVYY421 pKa = 10.28PNPTQEE427 pKa = 3.43SWTIKK432 pKa = 10.09AQNTKK437 pKa = 8.11ITNIKK442 pKa = 10.18VYY444 pKa = 10.67DD445 pKa = 3.78VLGKK449 pKa = 10.14NVMMLSPNTIEE460 pKa = 4.17TKK462 pKa = 10.11IDD464 pKa = 3.31GASLMKK470 pKa = 10.58GIYY473 pKa = 8.37FAKK476 pKa = 10.07IEE478 pKa = 4.47TVSGTSSVKK487 pKa = 10.21LVKK490 pKa = 10.26QQ491 pKa = 3.39
Molecular weight: 51.97 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H9BXK1|A0A1H9BXK1_9FLAO Magnesium transporter MgtE OS=Hyunsoonleella jejuensis OX=419940 GN=SAMN05421824_0742 PE=3 SV=1
MM1 pKa = 7.84PSGKK5 pKa = 9.32KK6 pKa = 9.59RR7 pKa = 11.84KK8 pKa = 7.05RR9 pKa = 11.84HH10 pKa = 5.11KK11 pKa = 10.59VATHH15 pKa = 5.21KK16 pKa = 10.3RR17 pKa = 11.84KK18 pKa = 9.62KK19 pKa = 9.04RR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84ANRR25 pKa = 11.84HH26 pKa = 4.77KK27 pKa = 10.58KK28 pKa = 9.8KK29 pKa = 10.51KK30 pKa = 10.04
MM1 pKa = 7.84PSGKK5 pKa = 9.32KK6 pKa = 9.59RR7 pKa = 11.84KK8 pKa = 7.05RR9 pKa = 11.84HH10 pKa = 5.11KK11 pKa = 10.59VATHH15 pKa = 5.21KK16 pKa = 10.3RR17 pKa = 11.84KK18 pKa = 9.62KK19 pKa = 9.04RR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84ANRR25 pKa = 11.84HH26 pKa = 4.77KK27 pKa = 10.58KK28 pKa = 9.8KK29 pKa = 10.51KK30 pKa = 10.04
Molecular weight: 3.76 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1056956 |
30 |
3421 |
359.6 |
40.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.421 ± 0.04 | 0.804 ± 0.015 |
5.809 ± 0.036 | 6.491 ± 0.048 |
5.224 ± 0.036 | 6.418 ± 0.047 |
1.817 ± 0.022 | 7.893 ± 0.042 |
7.766 ± 0.072 | 9.091 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.109 ± 0.024 | 6.368 ± 0.048 |
3.421 ± 0.027 | 3.194 ± 0.022 |
3.457 ± 0.031 | 6.375 ± 0.041 |
5.862 ± 0.055 | 6.326 ± 0.033 |
1.079 ± 0.02 | 4.076 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
