
Planaria asexual strain-specific virus-like element type 1
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.66
Get precalculated fractions of proteins
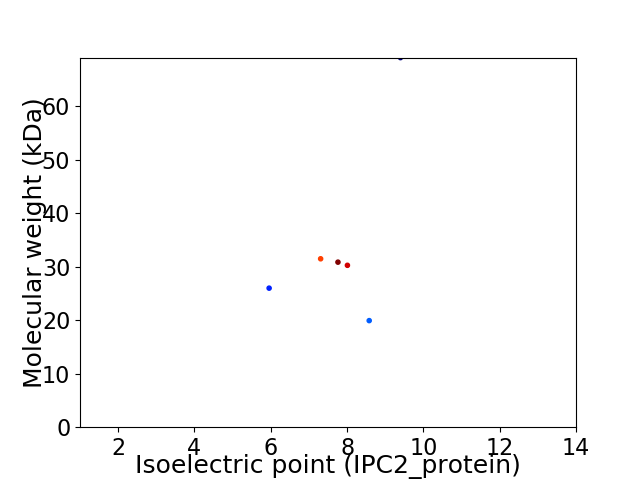
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q91S74|Q91S74_9VIRU ORF1S OS=Planaria asexual strain-specific virus-like element type 1 OX=159252 GN=ORF1S PE=4 SV=1
MM1 pKa = 7.39NLIQAISDD9 pKa = 4.01DD10 pKa = 4.0EE11 pKa = 4.28LQLFIINAMDD21 pKa = 4.45HH22 pKa = 7.01IIRR25 pKa = 11.84HH26 pKa = 5.25SVFEE30 pKa = 4.39YY31 pKa = 10.74NDD33 pKa = 2.98SHH35 pKa = 7.92RR36 pKa = 11.84NVIEE40 pKa = 5.1KK41 pKa = 8.96MLQYY45 pKa = 11.22AGVKK49 pKa = 9.9KK50 pKa = 10.57EE51 pKa = 3.92NEE53 pKa = 4.14DD54 pKa = 3.6YY55 pKa = 10.42EE56 pKa = 4.9FKK58 pKa = 10.66YY59 pKa = 9.62KK60 pKa = 10.09TMAIISILEE69 pKa = 3.94MQAGKK74 pKa = 8.87TNVILMVGKK83 pKa = 8.83PNCGKK88 pKa = 10.56SLFCNVLCAGLKK100 pKa = 9.31TGAAHH105 pKa = 7.04SMNSRR110 pKa = 11.84SNFWLQDD117 pKa = 3.58LQGKK121 pKa = 9.56DD122 pKa = 2.58IYY124 pKa = 11.32VFEE127 pKa = 4.58EE128 pKa = 4.32FSMDD132 pKa = 3.45VFNANYY138 pKa = 9.67FKK140 pKa = 11.16ALLEE144 pKa = 4.33GSMHH148 pKa = 7.0VMVEE152 pKa = 4.23KK153 pKa = 10.96KK154 pKa = 10.56NMPSVHH160 pKa = 6.46LEE162 pKa = 3.65RR163 pKa = 11.84RR164 pKa = 11.84PVITTGNEE172 pKa = 4.17FPWKK176 pKa = 9.69LVSSTAEE183 pKa = 3.75AFRR186 pKa = 11.84EE187 pKa = 3.67RR188 pKa = 11.84SLIILTDD195 pKa = 3.41SQIPFSVSADD205 pKa = 3.49PNKK208 pKa = 10.78RR209 pKa = 11.84PDD211 pKa = 4.08LLLNCKK217 pKa = 9.84NLLYY221 pKa = 10.62EE222 pKa = 4.83SIKK225 pKa = 11.04NCC227 pKa = 4.49
MM1 pKa = 7.39NLIQAISDD9 pKa = 4.01DD10 pKa = 4.0EE11 pKa = 4.28LQLFIINAMDD21 pKa = 4.45HH22 pKa = 7.01IIRR25 pKa = 11.84HH26 pKa = 5.25SVFEE30 pKa = 4.39YY31 pKa = 10.74NDD33 pKa = 2.98SHH35 pKa = 7.92RR36 pKa = 11.84NVIEE40 pKa = 5.1KK41 pKa = 8.96MLQYY45 pKa = 11.22AGVKK49 pKa = 9.9KK50 pKa = 10.57EE51 pKa = 3.92NEE53 pKa = 4.14DD54 pKa = 3.6YY55 pKa = 10.42EE56 pKa = 4.9FKK58 pKa = 10.66YY59 pKa = 9.62KK60 pKa = 10.09TMAIISILEE69 pKa = 3.94MQAGKK74 pKa = 8.87TNVILMVGKK83 pKa = 8.83PNCGKK88 pKa = 10.56SLFCNVLCAGLKK100 pKa = 9.31TGAAHH105 pKa = 7.04SMNSRR110 pKa = 11.84SNFWLQDD117 pKa = 3.58LQGKK121 pKa = 9.56DD122 pKa = 2.58IYY124 pKa = 11.32VFEE127 pKa = 4.58EE128 pKa = 4.32FSMDD132 pKa = 3.45VFNANYY138 pKa = 9.67FKK140 pKa = 11.16ALLEE144 pKa = 4.33GSMHH148 pKa = 7.0VMVEE152 pKa = 4.23KK153 pKa = 10.96KK154 pKa = 10.56NMPSVHH160 pKa = 6.46LEE162 pKa = 3.65RR163 pKa = 11.84RR164 pKa = 11.84PVITTGNEE172 pKa = 4.17FPWKK176 pKa = 9.69LVSSTAEE183 pKa = 3.75AFRR186 pKa = 11.84EE187 pKa = 3.67RR188 pKa = 11.84SLIILTDD195 pKa = 3.41SQIPFSVSADD205 pKa = 3.49PNKK208 pKa = 10.78RR209 pKa = 11.84PDD211 pKa = 4.08LLLNCKK217 pKa = 9.84NLLYY221 pKa = 10.62EE222 pKa = 4.83SIKK225 pKa = 11.04NCC227 pKa = 4.49
Molecular weight: 25.99 kDa
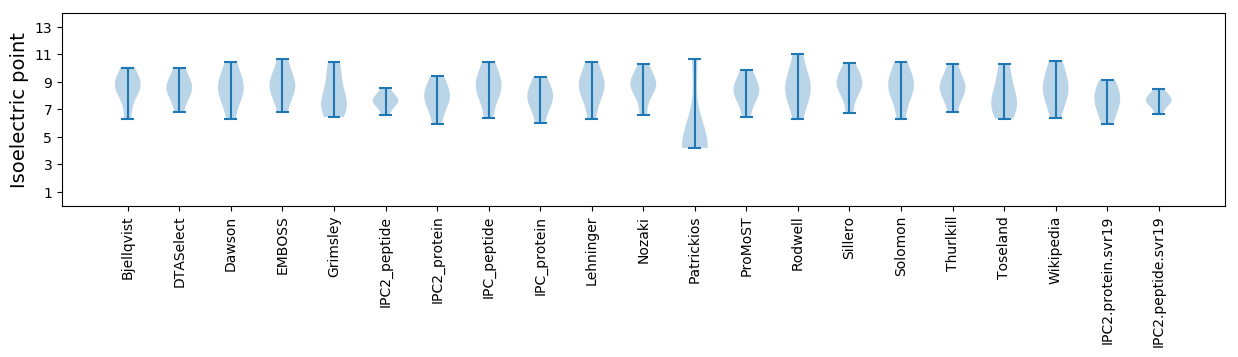
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q91S72|Q91S72_9VIRU ORF1L OS=Planaria asexual strain-specific virus-like element type 1 OX=159252 GN=ORF1L PE=4 SV=1
MM1 pKa = 7.83GIRR4 pKa = 11.84RR5 pKa = 11.84KK6 pKa = 10.08KK7 pKa = 7.55VTKK10 pKa = 9.48TVTPKK15 pKa = 9.82RR16 pKa = 11.84TTQHH20 pKa = 5.48IQNPAYY26 pKa = 10.28SGTPGGPQAASTPLRR41 pKa = 11.84NTPTRR46 pKa = 11.84TPPTKK51 pKa = 8.5TTPIRR56 pKa = 11.84NRR58 pKa = 11.84LRR60 pKa = 11.84SRR62 pKa = 11.84VVGRR66 pKa = 11.84NGQAVDD72 pKa = 4.31RR73 pKa = 11.84PLLSYY78 pKa = 10.32RR79 pKa = 11.84GRR81 pKa = 11.84GANFGIMHH89 pKa = 7.18NSPTNPRR96 pKa = 11.84NIMRR100 pKa = 11.84MQPRR104 pKa = 11.84VMVEE108 pKa = 3.85NILNLSNYY116 pKa = 9.69RR117 pKa = 11.84EE118 pKa = 4.25TSFGTPTQTPRR129 pKa = 11.84QTPQVTPRR137 pKa = 11.84NSPTQPPIQPPNPEE151 pKa = 4.06IPRR154 pKa = 11.84PSQPIINKK162 pKa = 8.12NTDD165 pKa = 2.83GTGPGPSGVQYY176 pKa = 10.71TKK178 pKa = 11.29DD179 pKa = 3.23NTILDD184 pKa = 3.89EE185 pKa = 4.84SSILKK190 pKa = 10.12DD191 pKa = 3.18KK192 pKa = 10.43KK193 pKa = 10.31RR194 pKa = 11.84NYY196 pKa = 9.26NLRR199 pKa = 11.84RR200 pKa = 11.84RR201 pKa = 11.84LISKK205 pKa = 8.42FKK207 pKa = 11.14NNNNKK212 pKa = 10.13NISTKK217 pKa = 10.89SNIPLIPMTSFKK229 pKa = 10.86NKK231 pKa = 9.26PQILPKK237 pKa = 9.45PQPKK241 pKa = 10.42GILKK245 pKa = 10.13PNIIPKK251 pKa = 9.96IKK253 pKa = 9.35PAYY256 pKa = 7.94PPKK259 pKa = 10.63HH260 pKa = 6.48FITNKK265 pKa = 9.37PPPIRR270 pKa = 11.84YY271 pKa = 8.9NPSSKK276 pKa = 10.22ILGWGTDD283 pKa = 2.97KK284 pKa = 11.14GVKK287 pKa = 9.2IKK289 pKa = 10.9SGVGGILGIGKK300 pKa = 7.74EE301 pKa = 4.34TTKK304 pKa = 10.62LGHH307 pKa = 6.67FIKK310 pKa = 10.05GTAKK314 pKa = 10.19AGLIIGLGLTLGQVLYY330 pKa = 9.15ATGQYY335 pKa = 8.95FQQARR340 pKa = 11.84IDD342 pKa = 4.28KK343 pKa = 9.36MAFDD347 pKa = 6.18DD348 pKa = 3.49IHH350 pKa = 8.12NEE352 pKa = 3.47IKK354 pKa = 9.88TVFPRR359 pKa = 11.84LVLMNRR365 pKa = 11.84YY366 pKa = 8.45VKK368 pKa = 10.31QFQNFKK374 pKa = 10.48FKK376 pKa = 10.94HH377 pKa = 5.75NPFKK381 pKa = 10.67DD382 pKa = 4.01TIPKK386 pKa = 9.95AIQRR390 pKa = 11.84LADD393 pKa = 3.2QALNIEE399 pKa = 4.38RR400 pKa = 11.84LKK402 pKa = 10.58IAHH405 pKa = 6.55KK406 pKa = 10.44NSYY409 pKa = 9.71FGVRR413 pKa = 11.84IPQEE417 pKa = 4.16TKK419 pKa = 10.22DD420 pKa = 3.85KK421 pKa = 10.96YY422 pKa = 10.79VNLYY426 pKa = 7.32TQILSVVNEE435 pKa = 3.95VNKK438 pKa = 9.22TVKK441 pKa = 10.24PLAIEE446 pKa = 4.32LNNLTYY452 pKa = 10.1EE453 pKa = 4.23EE454 pKa = 4.51CVEE457 pKa = 3.42IHH459 pKa = 6.21RR460 pKa = 11.84RR461 pKa = 11.84KK462 pKa = 7.6TEE464 pKa = 3.62KK465 pKa = 9.78MKK467 pKa = 11.01QEE469 pKa = 3.92MEE471 pKa = 4.2DD472 pKa = 3.87AEE474 pKa = 4.52EE475 pKa = 4.32EE476 pKa = 4.26DD477 pKa = 3.85PSKK480 pKa = 10.8IFPGPGKK487 pKa = 10.34IPLDD491 pKa = 3.86EE492 pKa = 5.0DD493 pKa = 3.3MKK495 pKa = 11.22KK496 pKa = 10.8YY497 pKa = 10.57INPHH501 pKa = 4.64TNMSYY506 pKa = 9.87TEE508 pKa = 4.43EE509 pKa = 4.05EE510 pKa = 4.56WSNLPADD517 pKa = 4.8IIQKK521 pKa = 9.74IEE523 pKa = 3.82NGEE526 pKa = 4.15QVSVGPQTSGDD537 pKa = 3.44GGIGVAEE544 pKa = 4.31NQPVPPPSISEE555 pKa = 4.02RR556 pKa = 11.84EE557 pKa = 3.88RR558 pKa = 11.84EE559 pKa = 4.3VNEE562 pKa = 4.07EE563 pKa = 3.77EE564 pKa = 5.14AKK566 pKa = 10.3QDD568 pKa = 3.65EE569 pKa = 4.45NNITSKK575 pKa = 10.93SAICPHH581 pKa = 5.98GRR583 pKa = 11.84CRR585 pKa = 11.84QILVEE590 pKa = 4.01KK591 pKa = 10.11AVRR594 pKa = 11.84STASCRR600 pKa = 11.84APAFVQIHH608 pKa = 4.55HH609 pKa = 6.01QKK611 pKa = 9.1RR612 pKa = 11.84TFF614 pKa = 3.1
MM1 pKa = 7.83GIRR4 pKa = 11.84RR5 pKa = 11.84KK6 pKa = 10.08KK7 pKa = 7.55VTKK10 pKa = 9.48TVTPKK15 pKa = 9.82RR16 pKa = 11.84TTQHH20 pKa = 5.48IQNPAYY26 pKa = 10.28SGTPGGPQAASTPLRR41 pKa = 11.84NTPTRR46 pKa = 11.84TPPTKK51 pKa = 8.5TTPIRR56 pKa = 11.84NRR58 pKa = 11.84LRR60 pKa = 11.84SRR62 pKa = 11.84VVGRR66 pKa = 11.84NGQAVDD72 pKa = 4.31RR73 pKa = 11.84PLLSYY78 pKa = 10.32RR79 pKa = 11.84GRR81 pKa = 11.84GANFGIMHH89 pKa = 7.18NSPTNPRR96 pKa = 11.84NIMRR100 pKa = 11.84MQPRR104 pKa = 11.84VMVEE108 pKa = 3.85NILNLSNYY116 pKa = 9.69RR117 pKa = 11.84EE118 pKa = 4.25TSFGTPTQTPRR129 pKa = 11.84QTPQVTPRR137 pKa = 11.84NSPTQPPIQPPNPEE151 pKa = 4.06IPRR154 pKa = 11.84PSQPIINKK162 pKa = 8.12NTDD165 pKa = 2.83GTGPGPSGVQYY176 pKa = 10.71TKK178 pKa = 11.29DD179 pKa = 3.23NTILDD184 pKa = 3.89EE185 pKa = 4.84SSILKK190 pKa = 10.12DD191 pKa = 3.18KK192 pKa = 10.43KK193 pKa = 10.31RR194 pKa = 11.84NYY196 pKa = 9.26NLRR199 pKa = 11.84RR200 pKa = 11.84RR201 pKa = 11.84LISKK205 pKa = 8.42FKK207 pKa = 11.14NNNNKK212 pKa = 10.13NISTKK217 pKa = 10.89SNIPLIPMTSFKK229 pKa = 10.86NKK231 pKa = 9.26PQILPKK237 pKa = 9.45PQPKK241 pKa = 10.42GILKK245 pKa = 10.13PNIIPKK251 pKa = 9.96IKK253 pKa = 9.35PAYY256 pKa = 7.94PPKK259 pKa = 10.63HH260 pKa = 6.48FITNKK265 pKa = 9.37PPPIRR270 pKa = 11.84YY271 pKa = 8.9NPSSKK276 pKa = 10.22ILGWGTDD283 pKa = 2.97KK284 pKa = 11.14GVKK287 pKa = 9.2IKK289 pKa = 10.9SGVGGILGIGKK300 pKa = 7.74EE301 pKa = 4.34TTKK304 pKa = 10.62LGHH307 pKa = 6.67FIKK310 pKa = 10.05GTAKK314 pKa = 10.19AGLIIGLGLTLGQVLYY330 pKa = 9.15ATGQYY335 pKa = 8.95FQQARR340 pKa = 11.84IDD342 pKa = 4.28KK343 pKa = 9.36MAFDD347 pKa = 6.18DD348 pKa = 3.49IHH350 pKa = 8.12NEE352 pKa = 3.47IKK354 pKa = 9.88TVFPRR359 pKa = 11.84LVLMNRR365 pKa = 11.84YY366 pKa = 8.45VKK368 pKa = 10.31QFQNFKK374 pKa = 10.48FKK376 pKa = 10.94HH377 pKa = 5.75NPFKK381 pKa = 10.67DD382 pKa = 4.01TIPKK386 pKa = 9.95AIQRR390 pKa = 11.84LADD393 pKa = 3.2QALNIEE399 pKa = 4.38RR400 pKa = 11.84LKK402 pKa = 10.58IAHH405 pKa = 6.55KK406 pKa = 10.44NSYY409 pKa = 9.71FGVRR413 pKa = 11.84IPQEE417 pKa = 4.16TKK419 pKa = 10.22DD420 pKa = 3.85KK421 pKa = 10.96YY422 pKa = 10.79VNLYY426 pKa = 7.32TQILSVVNEE435 pKa = 3.95VNKK438 pKa = 9.22TVKK441 pKa = 10.24PLAIEE446 pKa = 4.32LNNLTYY452 pKa = 10.1EE453 pKa = 4.23EE454 pKa = 4.51CVEE457 pKa = 3.42IHH459 pKa = 6.21RR460 pKa = 11.84RR461 pKa = 11.84KK462 pKa = 7.6TEE464 pKa = 3.62KK465 pKa = 9.78MKK467 pKa = 11.01QEE469 pKa = 3.92MEE471 pKa = 4.2DD472 pKa = 3.87AEE474 pKa = 4.52EE475 pKa = 4.32EE476 pKa = 4.26DD477 pKa = 3.85PSKK480 pKa = 10.8IFPGPGKK487 pKa = 10.34IPLDD491 pKa = 3.86EE492 pKa = 5.0DD493 pKa = 3.3MKK495 pKa = 11.22KK496 pKa = 10.8YY497 pKa = 10.57INPHH501 pKa = 4.64TNMSYY506 pKa = 9.87TEE508 pKa = 4.43EE509 pKa = 4.05EE510 pKa = 4.56WSNLPADD517 pKa = 4.8IIQKK521 pKa = 9.74IEE523 pKa = 3.82NGEE526 pKa = 4.15QVSVGPQTSGDD537 pKa = 3.44GGIGVAEE544 pKa = 4.31NQPVPPPSISEE555 pKa = 4.02RR556 pKa = 11.84EE557 pKa = 3.88RR558 pKa = 11.84EE559 pKa = 4.3VNEE562 pKa = 4.07EE563 pKa = 3.77EE564 pKa = 5.14AKK566 pKa = 10.3QDD568 pKa = 3.65EE569 pKa = 4.45NNITSKK575 pKa = 10.93SAICPHH581 pKa = 5.98GRR583 pKa = 11.84CRR585 pKa = 11.84QILVEE590 pKa = 4.01KK591 pKa = 10.11AVRR594 pKa = 11.84STASCRR600 pKa = 11.84APAFVQIHH608 pKa = 4.55HH609 pKa = 6.01QKK611 pKa = 9.1RR612 pKa = 11.84TFF614 pKa = 3.1
Molecular weight: 69.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1807 |
165 |
614 |
301.2 |
34.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.317 ± 0.311 | 1.328 ± 0.346 |
4.483 ± 0.564 | 5.589 ± 0.527 |
4.815 ± 0.904 | 5.589 ± 0.727 |
2.546 ± 0.264 | 6.807 ± 0.808 |
7.305 ± 0.83 | 7.914 ± 1.178 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.21 ± 0.56 | 6.309 ± 0.708 |
6.53 ± 1.369 | 4.87 ± 0.533 |
5.811 ± 0.433 | 5.811 ± 0.503 |
6.032 ± 0.827 | 5.921 ± 0.802 |
1.051 ± 0.374 | 3.763 ± 0.659 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
