
Alces alces faeces associated genomovirus MP157
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus alces2
Average proteome isoelectric point is 7.32
Get precalculated fractions of proteins
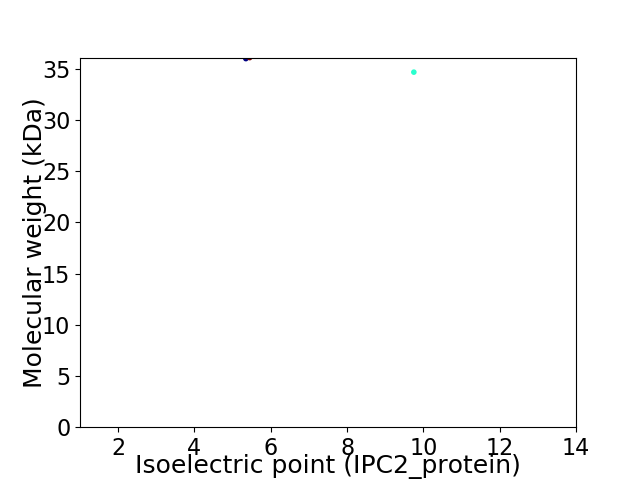
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Z5CJA5|A0A2Z5CJA5_9VIRU Replication associated protein OS=Alces alces faeces associated genomovirus MP157 OX=2219113 PE=4 SV=1
MM1 pKa = 7.72PFDD4 pKa = 3.32FHH6 pKa = 8.54SRR8 pKa = 11.84YY9 pKa = 10.7GLFTYY14 pKa = 10.71AHH16 pKa = 6.52VDD18 pKa = 3.44EE19 pKa = 5.93LDD21 pKa = 3.6PFCIVEE27 pKa = 4.48HH28 pKa = 6.37FSSLGGEE35 pKa = 4.44CIVAAEE41 pKa = 5.01RR42 pKa = 11.84YY43 pKa = 6.0PTGPGVHH50 pKa = 5.25YY51 pKa = 10.57HH52 pKa = 5.67VFTDD56 pKa = 3.96FGRR59 pKa = 11.84KK60 pKa = 7.93FRR62 pKa = 11.84SRR64 pKa = 11.84RR65 pKa = 11.84VDD67 pKa = 3.45CFDD70 pKa = 3.29VAGFHH75 pKa = 6.56PNIVASRR82 pKa = 11.84GTPEE86 pKa = 4.14AGYY89 pKa = 10.62DD90 pKa = 3.63YY91 pKa = 10.68AIKK94 pKa = 10.92DD95 pKa = 3.59GDD97 pKa = 4.01VVAGGLEE104 pKa = 4.06RR105 pKa = 11.84PSGVEE110 pKa = 3.23PDD112 pKa = 3.3TRR114 pKa = 11.84EE115 pKa = 4.4TKK117 pKa = 7.53WHH119 pKa = 7.25RR120 pKa = 11.84ITSAATRR127 pKa = 11.84EE128 pKa = 4.32EE129 pKa = 4.38FFHH132 pKa = 7.27LLRR135 pKa = 11.84TLDD138 pKa = 3.82PKK140 pKa = 11.34SLVTNHH146 pKa = 6.88RR147 pKa = 11.84SCIAFADD154 pKa = 3.34WQYY157 pKa = 11.12RR158 pKa = 11.84VEE160 pKa = 4.15PTPYY164 pKa = 8.89ATPNGVFNLAEE175 pKa = 4.28YY176 pKa = 11.1GDD178 pKa = 4.3LSEE181 pKa = 4.87SRR183 pKa = 11.84SLVLFGDD190 pKa = 3.55TRR192 pKa = 11.84LGKK195 pKa = 7.65TVWSRR200 pKa = 11.84SLGPHH205 pKa = 6.34VYY207 pKa = 10.15FCGLYY212 pKa = 10.18SYY214 pKa = 11.49AEE216 pKa = 4.04AVKK219 pKa = 10.8APDD222 pKa = 2.58VDD224 pKa = 3.94YY225 pKa = 11.48AVFDD229 pKa = 5.38DD230 pKa = 3.82IQGGIKK236 pKa = 10.07FFPAFKK242 pKa = 10.48NWLGCQMEE250 pKa = 4.38FQVKK254 pKa = 9.64GLYY257 pKa = 9.72RR258 pKa = 11.84DD259 pKa = 3.68PQLLKK264 pKa = 9.92WGKK267 pKa = 9.04PSIWVANTDD276 pKa = 3.34PRR278 pKa = 11.84HH279 pKa = 6.79DD280 pKa = 3.88MTHH283 pKa = 7.45DD284 pKa = 4.47DD285 pKa = 4.22IKK287 pKa = 10.95WMEE290 pKa = 4.11GNCIFVEE297 pKa = 4.19IVSAIFHH304 pKa = 6.95ASTGYY309 pKa = 8.9PACGEE314 pKa = 4.28TAEE317 pKa = 4.59SPP319 pKa = 3.6
MM1 pKa = 7.72PFDD4 pKa = 3.32FHH6 pKa = 8.54SRR8 pKa = 11.84YY9 pKa = 10.7GLFTYY14 pKa = 10.71AHH16 pKa = 6.52VDD18 pKa = 3.44EE19 pKa = 5.93LDD21 pKa = 3.6PFCIVEE27 pKa = 4.48HH28 pKa = 6.37FSSLGGEE35 pKa = 4.44CIVAAEE41 pKa = 5.01RR42 pKa = 11.84YY43 pKa = 6.0PTGPGVHH50 pKa = 5.25YY51 pKa = 10.57HH52 pKa = 5.67VFTDD56 pKa = 3.96FGRR59 pKa = 11.84KK60 pKa = 7.93FRR62 pKa = 11.84SRR64 pKa = 11.84RR65 pKa = 11.84VDD67 pKa = 3.45CFDD70 pKa = 3.29VAGFHH75 pKa = 6.56PNIVASRR82 pKa = 11.84GTPEE86 pKa = 4.14AGYY89 pKa = 10.62DD90 pKa = 3.63YY91 pKa = 10.68AIKK94 pKa = 10.92DD95 pKa = 3.59GDD97 pKa = 4.01VVAGGLEE104 pKa = 4.06RR105 pKa = 11.84PSGVEE110 pKa = 3.23PDD112 pKa = 3.3TRR114 pKa = 11.84EE115 pKa = 4.4TKK117 pKa = 7.53WHH119 pKa = 7.25RR120 pKa = 11.84ITSAATRR127 pKa = 11.84EE128 pKa = 4.32EE129 pKa = 4.38FFHH132 pKa = 7.27LLRR135 pKa = 11.84TLDD138 pKa = 3.82PKK140 pKa = 11.34SLVTNHH146 pKa = 6.88RR147 pKa = 11.84SCIAFADD154 pKa = 3.34WQYY157 pKa = 11.12RR158 pKa = 11.84VEE160 pKa = 4.15PTPYY164 pKa = 8.89ATPNGVFNLAEE175 pKa = 4.28YY176 pKa = 11.1GDD178 pKa = 4.3LSEE181 pKa = 4.87SRR183 pKa = 11.84SLVLFGDD190 pKa = 3.55TRR192 pKa = 11.84LGKK195 pKa = 7.65TVWSRR200 pKa = 11.84SLGPHH205 pKa = 6.34VYY207 pKa = 10.15FCGLYY212 pKa = 10.18SYY214 pKa = 11.49AEE216 pKa = 4.04AVKK219 pKa = 10.8APDD222 pKa = 2.58VDD224 pKa = 3.94YY225 pKa = 11.48AVFDD229 pKa = 5.38DD230 pKa = 3.82IQGGIKK236 pKa = 10.07FFPAFKK242 pKa = 10.48NWLGCQMEE250 pKa = 4.38FQVKK254 pKa = 9.64GLYY257 pKa = 9.72RR258 pKa = 11.84DD259 pKa = 3.68PQLLKK264 pKa = 9.92WGKK267 pKa = 9.04PSIWVANTDD276 pKa = 3.34PRR278 pKa = 11.84HH279 pKa = 6.79DD280 pKa = 3.88MTHH283 pKa = 7.45DD284 pKa = 4.47DD285 pKa = 4.22IKK287 pKa = 10.95WMEE290 pKa = 4.11GNCIFVEE297 pKa = 4.19IVSAIFHH304 pKa = 6.95ASTGYY309 pKa = 8.9PACGEE314 pKa = 4.28TAEE317 pKa = 4.59SPP319 pKa = 3.6
Molecular weight: 36.0 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z5CJA5|A0A2Z5CJA5_9VIRU Replication associated protein OS=Alces alces faeces associated genomovirus MP157 OX=2219113 PE=4 SV=1
MM1 pKa = 7.74GYY3 pKa = 10.68AKK5 pKa = 10.22RR6 pKa = 11.84RR7 pKa = 11.84STSSRR12 pKa = 11.84RR13 pKa = 11.84TTRR16 pKa = 11.84GPYY19 pKa = 9.47RR20 pKa = 11.84RR21 pKa = 11.84YY22 pKa = 6.46TAKK25 pKa = 10.25RR26 pKa = 11.84RR27 pKa = 11.84SLGKK31 pKa = 9.93AKK33 pKa = 10.13RR34 pKa = 11.84PYY36 pKa = 9.85RR37 pKa = 11.84RR38 pKa = 11.84TYY40 pKa = 9.9RR41 pKa = 11.84KK42 pKa = 10.15KK43 pKa = 10.04PMTKK47 pKa = 9.69RR48 pKa = 11.84RR49 pKa = 11.84ILNATSTKK57 pKa = 10.49KK58 pKa = 10.11KK59 pKa = 10.87DD60 pKa = 3.43NMLISTNTTAANPIGGATYY79 pKa = 7.59TTNPAILIGAQTFDD93 pKa = 3.45YY94 pKa = 10.66VFPFLCTYY102 pKa = 10.94RR103 pKa = 11.84DD104 pKa = 3.53FTTRR108 pKa = 11.84TGGSHH113 pKa = 6.54GFASDD118 pKa = 2.92TSTRR122 pKa = 11.84SASTVFMRR130 pKa = 11.84GYY132 pKa = 9.82KK133 pKa = 9.84EE134 pKa = 5.04CITIQTSDD142 pKa = 3.48GVPWLWRR149 pKa = 11.84RR150 pKa = 11.84ILISLKK156 pKa = 10.31GPLIAEE162 pKa = 4.26ISSVFNVAQEE172 pKa = 3.95NSAGFTRR179 pKa = 11.84VVNAANPGQRR189 pKa = 11.84ANVYY193 pKa = 10.57NLIFDD198 pKa = 4.65GTQDD202 pKa = 4.1ADD204 pKa = 3.54WNSPIDD210 pKa = 3.71AKK212 pKa = 10.88LDD214 pKa = 3.85RR215 pKa = 11.84NRR217 pKa = 11.84VTVMYY222 pKa = 10.26DD223 pKa = 3.01KK224 pKa = 10.93TITLASGNEE233 pKa = 3.85DD234 pKa = 3.48GMIRR238 pKa = 11.84KK239 pKa = 8.88YY240 pKa = 10.94NRR242 pKa = 11.84WHH244 pKa = 6.45GFNKK248 pKa = 10.02NFVYY252 pKa = 10.74DD253 pKa = 3.76DD254 pKa = 4.34DD255 pKa = 4.43KK256 pKa = 11.98AGGQEE261 pKa = 4.03NGSGYY266 pKa = 9.98HH267 pKa = 5.62AQGKK271 pKa = 10.22AGMGDD276 pKa = 3.65VYY278 pKa = 11.09VIDD281 pKa = 3.61IFRR284 pKa = 11.84PRR286 pKa = 11.84TGTTTATQLSLHH298 pKa = 5.61MQGTLYY304 pKa = 9.8WHH306 pKa = 6.96EE307 pKa = 4.2KK308 pKa = 9.34
MM1 pKa = 7.74GYY3 pKa = 10.68AKK5 pKa = 10.22RR6 pKa = 11.84RR7 pKa = 11.84STSSRR12 pKa = 11.84RR13 pKa = 11.84TTRR16 pKa = 11.84GPYY19 pKa = 9.47RR20 pKa = 11.84RR21 pKa = 11.84YY22 pKa = 6.46TAKK25 pKa = 10.25RR26 pKa = 11.84RR27 pKa = 11.84SLGKK31 pKa = 9.93AKK33 pKa = 10.13RR34 pKa = 11.84PYY36 pKa = 9.85RR37 pKa = 11.84RR38 pKa = 11.84TYY40 pKa = 9.9RR41 pKa = 11.84KK42 pKa = 10.15KK43 pKa = 10.04PMTKK47 pKa = 9.69RR48 pKa = 11.84RR49 pKa = 11.84ILNATSTKK57 pKa = 10.49KK58 pKa = 10.11KK59 pKa = 10.87DD60 pKa = 3.43NMLISTNTTAANPIGGATYY79 pKa = 7.59TTNPAILIGAQTFDD93 pKa = 3.45YY94 pKa = 10.66VFPFLCTYY102 pKa = 10.94RR103 pKa = 11.84DD104 pKa = 3.53FTTRR108 pKa = 11.84TGGSHH113 pKa = 6.54GFASDD118 pKa = 2.92TSTRR122 pKa = 11.84SASTVFMRR130 pKa = 11.84GYY132 pKa = 9.82KK133 pKa = 9.84EE134 pKa = 5.04CITIQTSDD142 pKa = 3.48GVPWLWRR149 pKa = 11.84RR150 pKa = 11.84ILISLKK156 pKa = 10.31GPLIAEE162 pKa = 4.26ISSVFNVAQEE172 pKa = 3.95NSAGFTRR179 pKa = 11.84VVNAANPGQRR189 pKa = 11.84ANVYY193 pKa = 10.57NLIFDD198 pKa = 4.65GTQDD202 pKa = 4.1ADD204 pKa = 3.54WNSPIDD210 pKa = 3.71AKK212 pKa = 10.88LDD214 pKa = 3.85RR215 pKa = 11.84NRR217 pKa = 11.84VTVMYY222 pKa = 10.26DD223 pKa = 3.01KK224 pKa = 10.93TITLASGNEE233 pKa = 3.85DD234 pKa = 3.48GMIRR238 pKa = 11.84KK239 pKa = 8.88YY240 pKa = 10.94NRR242 pKa = 11.84WHH244 pKa = 6.45GFNKK248 pKa = 10.02NFVYY252 pKa = 10.74DD253 pKa = 3.76DD254 pKa = 4.34DD255 pKa = 4.43KK256 pKa = 11.98AGGQEE261 pKa = 4.03NGSGYY266 pKa = 9.98HH267 pKa = 5.62AQGKK271 pKa = 10.22AGMGDD276 pKa = 3.65VYY278 pKa = 11.09VIDD281 pKa = 3.61IFRR284 pKa = 11.84PRR286 pKa = 11.84TGTTTATQLSLHH298 pKa = 5.61MQGTLYY304 pKa = 9.8WHH306 pKa = 6.96EE307 pKa = 4.2KK308 pKa = 9.34
Molecular weight: 34.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
627 |
308 |
319 |
313.5 |
35.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.656 ± 0.091 | 1.595 ± 0.63 |
6.38 ± 0.573 | 3.987 ± 1.36 |
5.582 ± 1.124 | 8.612 ± 0.103 |
2.871 ± 0.832 | 4.785 ± 0.49 |
4.944 ± 0.817 | 5.423 ± 0.368 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.914 ± 0.456 | 3.987 ± 1.238 |
4.944 ± 0.915 | 2.233 ± 0.46 |
7.656 ± 1.174 | 6.22 ± 0.182 |
8.453 ± 1.941 | 5.901 ± 1.12 |
1.914 ± 0.194 | 4.944 ± 0.167 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
