
Chiqui virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Spinareovirinae; unclassified Spinareovirinae
Average proteome isoelectric point is 5.94
Get precalculated fractions of proteins
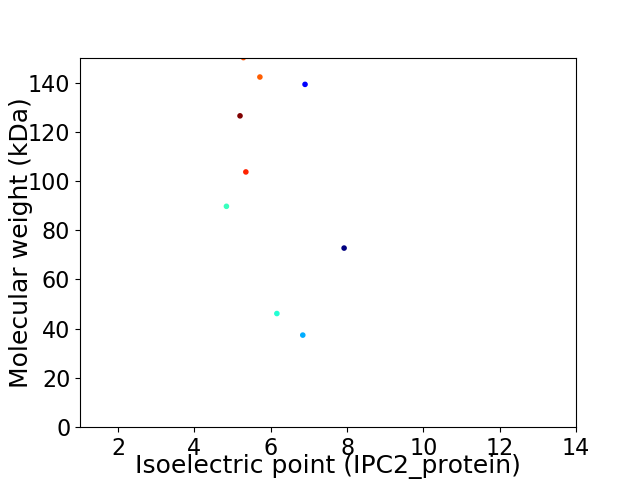
Virtual 2D-PAGE plot for 9 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Z4N706|A0A2Z4N706_9REOV P7 OS=Chiqui virus OX=2250219 GN=P7 PE=4 SV=1
MM1 pKa = 7.56ALIDD5 pKa = 4.08SEE7 pKa = 4.75ANQIRR12 pKa = 11.84IKK14 pKa = 9.92GANWQRR20 pKa = 11.84ITEE23 pKa = 4.29KK24 pKa = 10.86ARR26 pKa = 11.84LDD28 pKa = 3.6YY29 pKa = 10.19TQLSTILAQHH39 pKa = 6.43SSIQTASIIPIPEE52 pKa = 4.14DD53 pKa = 2.97QKK55 pKa = 10.17QTVKK59 pKa = 11.12NIVTVTDD66 pKa = 3.62TSGSADD72 pKa = 3.56NLNDD76 pKa = 4.15LLYY79 pKa = 10.79QLFIGSSKK87 pKa = 10.21PLEE90 pKa = 3.85IAKK93 pKa = 10.32KK94 pKa = 9.14IFRR97 pKa = 11.84IAATIVTQEE106 pKa = 3.84NSIITTNGYY115 pKa = 9.8FYY117 pKa = 10.88RR118 pKa = 11.84DD119 pKa = 3.68AASHH123 pKa = 5.38PMIRR127 pKa = 11.84QLEE130 pKa = 4.32IIEE133 pKa = 4.46RR134 pKa = 11.84NDD136 pKa = 4.2AINLFSKK143 pKa = 9.85YY144 pKa = 8.84YY145 pKa = 10.3RR146 pKa = 11.84WMEE149 pKa = 3.79LLLSNEE155 pKa = 4.16PLLVSDD161 pKa = 4.12QDD163 pKa = 3.72VGALLNVINVPTTVDD178 pKa = 3.52GQVVSDD184 pKa = 3.54SRR186 pKa = 11.84YY187 pKa = 8.52VVSDD191 pKa = 2.86VMFPNGHH198 pKa = 6.76ISTTIDD204 pKa = 3.19EE205 pKa = 4.57NLLRR209 pKa = 11.84TQIQDD214 pKa = 3.29MPLFGYY220 pKa = 10.42FSDD223 pKa = 5.15IITDD227 pKa = 3.42NFNFGTHH234 pKa = 5.51RR235 pKa = 11.84QHH237 pKa = 8.14LDD239 pKa = 3.05VLRR242 pKa = 11.84LVLIYY247 pKa = 10.73AFRR250 pKa = 11.84RR251 pKa = 11.84WFTNAGLVTSGIQIGALSTFNRR273 pKa = 11.84QNALHH278 pKa = 6.66NGDD281 pKa = 4.08DD282 pKa = 3.81TDD284 pKa = 3.91IEE286 pKa = 4.76SKK288 pKa = 11.0GRR290 pKa = 11.84ILPSLFLTALLPSEE304 pKa = 4.9LLRR307 pKa = 11.84KK308 pKa = 9.25VKK310 pKa = 10.6NAGGEE315 pKa = 4.28TNLDD319 pKa = 3.78EE320 pKa = 5.9IIADD324 pKa = 3.7NLEE327 pKa = 4.02EE328 pKa = 4.06LMSRR332 pKa = 11.84IGKK335 pKa = 10.02LSVNTPTKK343 pKa = 9.25ITQSALDD350 pKa = 3.34KK351 pKa = 10.52MFNGLYY357 pKa = 10.11KK358 pKa = 10.38ISDD361 pKa = 3.77FGGKK365 pKa = 9.03NLSQGYY371 pKa = 7.92EE372 pKa = 4.01HH373 pKa = 7.56ASKK376 pKa = 10.81QSIRR380 pKa = 11.84FSDD383 pKa = 3.71YY384 pKa = 10.67TKK386 pKa = 10.23MGFSKK391 pKa = 10.82VAGLALTDD399 pKa = 3.94SGSATLPTKK408 pKa = 10.26DD409 pKa = 4.72APDD412 pKa = 3.94PSIGEE417 pKa = 3.9YY418 pKa = 9.05YY419 pKa = 10.54ARR421 pKa = 11.84HH422 pKa = 5.06KK423 pKa = 9.96TEE425 pKa = 4.18LLQLIANEE433 pKa = 3.98RR434 pKa = 11.84LSVDD438 pKa = 3.45EE439 pKa = 4.86LSVIFGGKK447 pKa = 9.91LISTDD452 pKa = 3.05QKK454 pKa = 10.99LNQIFKK460 pKa = 10.16TMLMPSNTVTLVDD473 pKa = 3.61SSKK476 pKa = 11.05VSFPTLINQCITQNGIDD493 pKa = 4.89KK494 pKa = 10.59LPKK497 pKa = 9.9DD498 pKa = 4.25PKK500 pKa = 10.88VDD502 pKa = 3.12IPTLIRR508 pKa = 11.84TNWDD512 pKa = 2.57ISAEE516 pKa = 4.27TISEE520 pKa = 4.44LEE522 pKa = 3.98LSDD525 pKa = 3.62VGVEE529 pKa = 4.06GGYY532 pKa = 10.66DD533 pKa = 3.42YY534 pKa = 11.96SKK536 pKa = 11.05LFKK539 pKa = 10.57NKK541 pKa = 9.41PFTGYY546 pKa = 10.63DD547 pKa = 3.85FSQGGIKK554 pKa = 10.45LGTFGTTWYY563 pKa = 8.72TDD565 pKa = 3.02EE566 pKa = 4.83GVRR569 pKa = 11.84QVALSNEE576 pKa = 4.33VVGADD581 pKa = 3.17MLMARR586 pKa = 11.84MLDD589 pKa = 3.63KK590 pKa = 10.63RR591 pKa = 11.84WNLPSEE597 pKa = 4.51PGVQRR602 pKa = 11.84TMLEE606 pKa = 3.89MMKK609 pKa = 10.91DD610 pKa = 3.55DD611 pKa = 4.03MASEE615 pKa = 4.46NPTLPTILAEE625 pKa = 4.17GQIVFLWSQFATIMEE640 pKa = 4.58HH641 pKa = 7.84DD642 pKa = 4.84DD643 pKa = 3.53WNDD646 pKa = 3.46DD647 pKa = 4.79DD648 pKa = 6.61VIQEE652 pKa = 4.1WKK654 pKa = 10.69DD655 pKa = 3.19KK656 pKa = 11.09GGSLGDD662 pKa = 3.48HH663 pKa = 6.48VNNIFEE669 pKa = 4.35VAIPMLSKK677 pKa = 11.2AKK679 pKa = 9.32MSTVGDD685 pKa = 3.75HH686 pKa = 6.93FSRR689 pKa = 11.84TIRR692 pKa = 11.84DD693 pKa = 3.29TDD695 pKa = 3.12GTYY698 pKa = 10.97AKK700 pKa = 9.73EE701 pKa = 3.79VSYY704 pKa = 11.26NILDD708 pKa = 5.6LITHH712 pKa = 6.29GTSQIEE718 pKa = 4.41GGSTFQQFIATEE730 pKa = 4.28PEE732 pKa = 4.45LGDD735 pKa = 3.02QGYY738 pKa = 10.35NGRR741 pKa = 11.84NLYY744 pKa = 10.6AYY746 pKa = 7.56FTGLVNDD753 pKa = 5.49NIPIDD758 pKa = 4.68KK759 pKa = 8.11ITKK762 pKa = 9.81VITPAKK768 pKa = 9.35LRR770 pKa = 11.84SIVVSDD776 pKa = 3.58STTVSIGDD784 pKa = 3.54NSEE787 pKa = 4.14PLHH790 pKa = 6.59KK791 pKa = 10.69VLEE794 pKa = 4.27SQIISDD800 pKa = 3.54NTTEE804 pKa = 4.13FNN806 pKa = 3.71
MM1 pKa = 7.56ALIDD5 pKa = 4.08SEE7 pKa = 4.75ANQIRR12 pKa = 11.84IKK14 pKa = 9.92GANWQRR20 pKa = 11.84ITEE23 pKa = 4.29KK24 pKa = 10.86ARR26 pKa = 11.84LDD28 pKa = 3.6YY29 pKa = 10.19TQLSTILAQHH39 pKa = 6.43SSIQTASIIPIPEE52 pKa = 4.14DD53 pKa = 2.97QKK55 pKa = 10.17QTVKK59 pKa = 11.12NIVTVTDD66 pKa = 3.62TSGSADD72 pKa = 3.56NLNDD76 pKa = 4.15LLYY79 pKa = 10.79QLFIGSSKK87 pKa = 10.21PLEE90 pKa = 3.85IAKK93 pKa = 10.32KK94 pKa = 9.14IFRR97 pKa = 11.84IAATIVTQEE106 pKa = 3.84NSIITTNGYY115 pKa = 9.8FYY117 pKa = 10.88RR118 pKa = 11.84DD119 pKa = 3.68AASHH123 pKa = 5.38PMIRR127 pKa = 11.84QLEE130 pKa = 4.32IIEE133 pKa = 4.46RR134 pKa = 11.84NDD136 pKa = 4.2AINLFSKK143 pKa = 9.85YY144 pKa = 8.84YY145 pKa = 10.3RR146 pKa = 11.84WMEE149 pKa = 3.79LLLSNEE155 pKa = 4.16PLLVSDD161 pKa = 4.12QDD163 pKa = 3.72VGALLNVINVPTTVDD178 pKa = 3.52GQVVSDD184 pKa = 3.54SRR186 pKa = 11.84YY187 pKa = 8.52VVSDD191 pKa = 2.86VMFPNGHH198 pKa = 6.76ISTTIDD204 pKa = 3.19EE205 pKa = 4.57NLLRR209 pKa = 11.84TQIQDD214 pKa = 3.29MPLFGYY220 pKa = 10.42FSDD223 pKa = 5.15IITDD227 pKa = 3.42NFNFGTHH234 pKa = 5.51RR235 pKa = 11.84QHH237 pKa = 8.14LDD239 pKa = 3.05VLRR242 pKa = 11.84LVLIYY247 pKa = 10.73AFRR250 pKa = 11.84RR251 pKa = 11.84WFTNAGLVTSGIQIGALSTFNRR273 pKa = 11.84QNALHH278 pKa = 6.66NGDD281 pKa = 4.08DD282 pKa = 3.81TDD284 pKa = 3.91IEE286 pKa = 4.76SKK288 pKa = 11.0GRR290 pKa = 11.84ILPSLFLTALLPSEE304 pKa = 4.9LLRR307 pKa = 11.84KK308 pKa = 9.25VKK310 pKa = 10.6NAGGEE315 pKa = 4.28TNLDD319 pKa = 3.78EE320 pKa = 5.9IIADD324 pKa = 3.7NLEE327 pKa = 4.02EE328 pKa = 4.06LMSRR332 pKa = 11.84IGKK335 pKa = 10.02LSVNTPTKK343 pKa = 9.25ITQSALDD350 pKa = 3.34KK351 pKa = 10.52MFNGLYY357 pKa = 10.11KK358 pKa = 10.38ISDD361 pKa = 3.77FGGKK365 pKa = 9.03NLSQGYY371 pKa = 7.92EE372 pKa = 4.01HH373 pKa = 7.56ASKK376 pKa = 10.81QSIRR380 pKa = 11.84FSDD383 pKa = 3.71YY384 pKa = 10.67TKK386 pKa = 10.23MGFSKK391 pKa = 10.82VAGLALTDD399 pKa = 3.94SGSATLPTKK408 pKa = 10.26DD409 pKa = 4.72APDD412 pKa = 3.94PSIGEE417 pKa = 3.9YY418 pKa = 9.05YY419 pKa = 10.54ARR421 pKa = 11.84HH422 pKa = 5.06KK423 pKa = 9.96TEE425 pKa = 4.18LLQLIANEE433 pKa = 3.98RR434 pKa = 11.84LSVDD438 pKa = 3.45EE439 pKa = 4.86LSVIFGGKK447 pKa = 9.91LISTDD452 pKa = 3.05QKK454 pKa = 10.99LNQIFKK460 pKa = 10.16TMLMPSNTVTLVDD473 pKa = 3.61SSKK476 pKa = 11.05VSFPTLINQCITQNGIDD493 pKa = 4.89KK494 pKa = 10.59LPKK497 pKa = 9.9DD498 pKa = 4.25PKK500 pKa = 10.88VDD502 pKa = 3.12IPTLIRR508 pKa = 11.84TNWDD512 pKa = 2.57ISAEE516 pKa = 4.27TISEE520 pKa = 4.44LEE522 pKa = 3.98LSDD525 pKa = 3.62VGVEE529 pKa = 4.06GGYY532 pKa = 10.66DD533 pKa = 3.42YY534 pKa = 11.96SKK536 pKa = 11.05LFKK539 pKa = 10.57NKK541 pKa = 9.41PFTGYY546 pKa = 10.63DD547 pKa = 3.85FSQGGIKK554 pKa = 10.45LGTFGTTWYY563 pKa = 8.72TDD565 pKa = 3.02EE566 pKa = 4.83GVRR569 pKa = 11.84QVALSNEE576 pKa = 4.33VVGADD581 pKa = 3.17MLMARR586 pKa = 11.84MLDD589 pKa = 3.63KK590 pKa = 10.63RR591 pKa = 11.84WNLPSEE597 pKa = 4.51PGVQRR602 pKa = 11.84TMLEE606 pKa = 3.89MMKK609 pKa = 10.91DD610 pKa = 3.55DD611 pKa = 4.03MASEE615 pKa = 4.46NPTLPTILAEE625 pKa = 4.17GQIVFLWSQFATIMEE640 pKa = 4.58HH641 pKa = 7.84DD642 pKa = 4.84DD643 pKa = 3.53WNDD646 pKa = 3.46DD647 pKa = 4.79DD648 pKa = 6.61VIQEE652 pKa = 4.1WKK654 pKa = 10.69DD655 pKa = 3.19KK656 pKa = 11.09GGSLGDD662 pKa = 3.48HH663 pKa = 6.48VNNIFEE669 pKa = 4.35VAIPMLSKK677 pKa = 11.2AKK679 pKa = 9.32MSTVGDD685 pKa = 3.75HH686 pKa = 6.93FSRR689 pKa = 11.84TIRR692 pKa = 11.84DD693 pKa = 3.29TDD695 pKa = 3.12GTYY698 pKa = 10.97AKK700 pKa = 9.73EE701 pKa = 3.79VSYY704 pKa = 11.26NILDD708 pKa = 5.6LITHH712 pKa = 6.29GTSQIEE718 pKa = 4.41GGSTFQQFIATEE730 pKa = 4.28PEE732 pKa = 4.45LGDD735 pKa = 3.02QGYY738 pKa = 10.35NGRR741 pKa = 11.84NLYY744 pKa = 10.6AYY746 pKa = 7.56FTGLVNDD753 pKa = 5.49NIPIDD758 pKa = 4.68KK759 pKa = 8.11ITKK762 pKa = 9.81VITPAKK768 pKa = 9.35LRR770 pKa = 11.84SIVVSDD776 pKa = 3.58STTVSIGDD784 pKa = 3.54NSEE787 pKa = 4.14PLHH790 pKa = 6.59KK791 pKa = 10.69VLEE794 pKa = 4.27SQIISDD800 pKa = 3.54NTTEE804 pKa = 4.13FNN806 pKa = 3.71
Molecular weight: 89.72 kDa
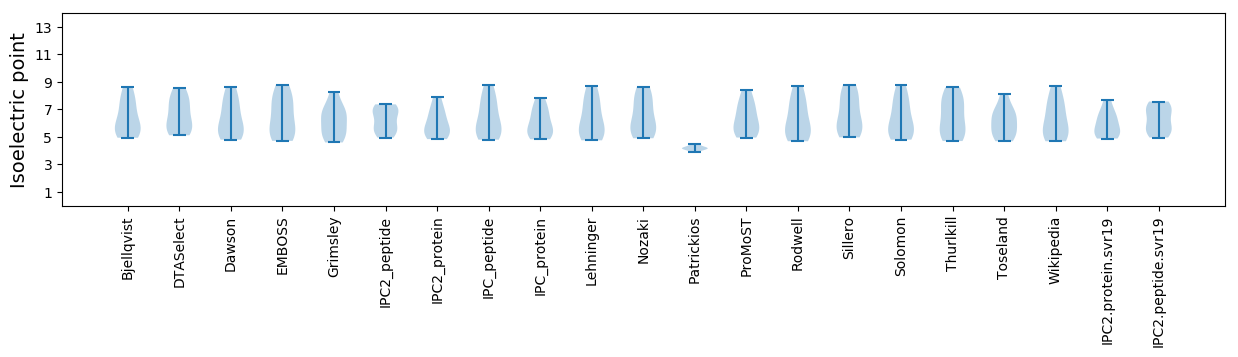
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z4N706|A0A2Z4N706_9REOV P7 OS=Chiqui virus OX=2250219 GN=P7 PE=4 SV=1
MM1 pKa = 7.75DD2 pKa = 4.72TLTGRR7 pKa = 11.84ITNDD11 pKa = 2.47SSAQEE16 pKa = 4.8DD17 pKa = 3.76LQIWPKK23 pKa = 9.21TLQNTRR29 pKa = 11.84EE30 pKa = 3.88ITYY33 pKa = 7.79TQQDD37 pKa = 3.38NQKK40 pKa = 8.26QQGRR44 pKa = 11.84KK45 pKa = 5.55VQKK48 pKa = 9.68SVKK51 pKa = 6.89THH53 pKa = 5.9SYY55 pKa = 8.88MLSTNEE61 pKa = 3.59VLQLHH66 pKa = 6.17EE67 pKa = 4.54MLIKK71 pKa = 10.01IYY73 pKa = 10.12NRR75 pKa = 11.84HH76 pKa = 6.42KK77 pKa = 10.48GTLPNPKK84 pKa = 10.16AIANLLNSQPVKK96 pKa = 10.69MMIEE100 pKa = 4.35SKK102 pKa = 11.03VKK104 pKa = 10.83LSDD107 pKa = 3.46DD108 pKa = 3.42QKK110 pKa = 11.52RR111 pKa = 11.84NLTEE115 pKa = 3.74IVTNTLANWTYY126 pKa = 11.37DD127 pKa = 3.3ADD129 pKa = 3.89HH130 pKa = 6.96FNMNKK135 pKa = 8.0QLRR138 pKa = 11.84RR139 pKa = 11.84AILNKK144 pKa = 9.5HH145 pKa = 6.18DD146 pKa = 4.66LSILVMDD153 pKa = 5.0NDD155 pKa = 4.66DD156 pKa = 4.39LQSATQTEE164 pKa = 4.38NTVTSITMALTIEE177 pKa = 4.28EE178 pKa = 4.44VAFLSNYY185 pKa = 10.11GKK187 pKa = 10.83GKK189 pKa = 10.12SCFVLYY195 pKa = 9.53TFNGPRR201 pKa = 11.84FAKK204 pKa = 10.15YY205 pKa = 8.79IHH207 pKa = 6.7PVYY210 pKa = 10.11TILSQVVPTISLSDD224 pKa = 3.65VKK226 pKa = 11.13NSLIDD231 pKa = 3.61TQHH234 pKa = 6.95PYY236 pKa = 10.05YY237 pKa = 10.34KK238 pKa = 10.66ARR240 pKa = 11.84FRR242 pKa = 11.84TLLTFDD248 pKa = 4.65EE249 pKa = 5.23FNDD252 pKa = 3.47NQSNYY257 pKa = 9.77RR258 pKa = 11.84KK259 pKa = 9.45VDD261 pKa = 3.21VVIITNSSEE270 pKa = 3.98NKK272 pKa = 9.8QKK274 pKa = 10.57VSSYY278 pKa = 7.37TRR280 pKa = 11.84KK281 pKa = 9.52GWKK284 pKa = 9.62FSQNKK289 pKa = 5.81QTTFYY294 pKa = 8.89WTMNGLTYY302 pKa = 10.61AGYY305 pKa = 9.25PLYY308 pKa = 10.64FMTKK312 pKa = 8.28ITPSDD317 pKa = 3.55YY318 pKa = 11.02RR319 pKa = 11.84DD320 pKa = 3.21ACVSYY325 pKa = 10.68GATGLVNNFKK335 pKa = 10.02IHH337 pKa = 6.34PNFYY341 pKa = 10.9LLLNSYY347 pKa = 10.31CRR349 pKa = 11.84INFKK353 pKa = 10.53PIVSALTRR361 pKa = 11.84SRR363 pKa = 11.84ISYY366 pKa = 9.71ISNKK370 pKa = 8.23MPVMPTEE377 pKa = 4.7DD378 pKa = 2.95MSAYY382 pKa = 9.73TKK384 pKa = 8.16MTMWLRR390 pKa = 11.84EE391 pKa = 3.54VDD393 pKa = 4.18RR394 pKa = 11.84IHH396 pKa = 7.09NGTEE400 pKa = 3.83SKK402 pKa = 10.58LRR404 pKa = 11.84NSSIIIIADD413 pKa = 3.27KK414 pKa = 11.16GSGKK418 pKa = 6.58TTCTKK423 pKa = 10.8YY424 pKa = 10.92LLQQLRR430 pKa = 11.84DD431 pKa = 3.69TSPSSKK437 pKa = 8.3TWGRR441 pKa = 11.84VDD443 pKa = 3.52SDD445 pKa = 5.29AFGKK449 pKa = 9.29WLSKK453 pKa = 9.74RR454 pKa = 11.84RR455 pKa = 11.84SMGMRR460 pKa = 11.84STDD463 pKa = 2.82ILLSNWQEE471 pKa = 3.7LDD473 pKa = 3.12ALQNDD478 pKa = 4.17EE479 pKa = 5.5SIPTYY484 pKa = 9.81FAHH487 pKa = 7.26EE488 pKa = 4.36MEE490 pKa = 4.74QMLLSAGMVEE500 pKa = 4.43YY501 pKa = 10.76GFDD504 pKa = 3.73PLSEE508 pKa = 4.08LRR510 pKa = 11.84ILDD513 pKa = 3.5EE514 pKa = 4.54FRR516 pKa = 11.84PIIYY520 pKa = 9.0TIINDD525 pKa = 3.8EE526 pKa = 4.41EE527 pKa = 3.95EE528 pKa = 4.29GLAAFYY534 pKa = 11.43NEE536 pKa = 4.0LDD538 pKa = 3.7KK539 pKa = 11.76FEE541 pKa = 5.47DD542 pKa = 3.94LPIGLMLEE550 pKa = 3.97SHH552 pKa = 6.79TDD554 pKa = 3.43VEE556 pKa = 4.23ISKK559 pKa = 9.3MFGTGHH565 pKa = 6.98IVTLDD570 pKa = 3.23APYY573 pKa = 10.33NAEE576 pKa = 3.75AAVADD581 pKa = 4.78RR582 pKa = 11.84KK583 pKa = 10.16RR584 pKa = 11.84SKK586 pKa = 10.5ISINVEE592 pKa = 3.64LMLFRR597 pKa = 11.84AWSRR601 pKa = 11.84LRR603 pKa = 11.84VSTYY607 pKa = 10.91PMLRR611 pKa = 11.84TQAFFDD617 pKa = 3.89TQLLAIASDD626 pKa = 3.67QEE628 pKa = 4.4TNPRR632 pKa = 3.56
MM1 pKa = 7.75DD2 pKa = 4.72TLTGRR7 pKa = 11.84ITNDD11 pKa = 2.47SSAQEE16 pKa = 4.8DD17 pKa = 3.76LQIWPKK23 pKa = 9.21TLQNTRR29 pKa = 11.84EE30 pKa = 3.88ITYY33 pKa = 7.79TQQDD37 pKa = 3.38NQKK40 pKa = 8.26QQGRR44 pKa = 11.84KK45 pKa = 5.55VQKK48 pKa = 9.68SVKK51 pKa = 6.89THH53 pKa = 5.9SYY55 pKa = 8.88MLSTNEE61 pKa = 3.59VLQLHH66 pKa = 6.17EE67 pKa = 4.54MLIKK71 pKa = 10.01IYY73 pKa = 10.12NRR75 pKa = 11.84HH76 pKa = 6.42KK77 pKa = 10.48GTLPNPKK84 pKa = 10.16AIANLLNSQPVKK96 pKa = 10.69MMIEE100 pKa = 4.35SKK102 pKa = 11.03VKK104 pKa = 10.83LSDD107 pKa = 3.46DD108 pKa = 3.42QKK110 pKa = 11.52RR111 pKa = 11.84NLTEE115 pKa = 3.74IVTNTLANWTYY126 pKa = 11.37DD127 pKa = 3.3ADD129 pKa = 3.89HH130 pKa = 6.96FNMNKK135 pKa = 8.0QLRR138 pKa = 11.84RR139 pKa = 11.84AILNKK144 pKa = 9.5HH145 pKa = 6.18DD146 pKa = 4.66LSILVMDD153 pKa = 5.0NDD155 pKa = 4.66DD156 pKa = 4.39LQSATQTEE164 pKa = 4.38NTVTSITMALTIEE177 pKa = 4.28EE178 pKa = 4.44VAFLSNYY185 pKa = 10.11GKK187 pKa = 10.83GKK189 pKa = 10.12SCFVLYY195 pKa = 9.53TFNGPRR201 pKa = 11.84FAKK204 pKa = 10.15YY205 pKa = 8.79IHH207 pKa = 6.7PVYY210 pKa = 10.11TILSQVVPTISLSDD224 pKa = 3.65VKK226 pKa = 11.13NSLIDD231 pKa = 3.61TQHH234 pKa = 6.95PYY236 pKa = 10.05YY237 pKa = 10.34KK238 pKa = 10.66ARR240 pKa = 11.84FRR242 pKa = 11.84TLLTFDD248 pKa = 4.65EE249 pKa = 5.23FNDD252 pKa = 3.47NQSNYY257 pKa = 9.77RR258 pKa = 11.84KK259 pKa = 9.45VDD261 pKa = 3.21VVIITNSSEE270 pKa = 3.98NKK272 pKa = 9.8QKK274 pKa = 10.57VSSYY278 pKa = 7.37TRR280 pKa = 11.84KK281 pKa = 9.52GWKK284 pKa = 9.62FSQNKK289 pKa = 5.81QTTFYY294 pKa = 8.89WTMNGLTYY302 pKa = 10.61AGYY305 pKa = 9.25PLYY308 pKa = 10.64FMTKK312 pKa = 8.28ITPSDD317 pKa = 3.55YY318 pKa = 11.02RR319 pKa = 11.84DD320 pKa = 3.21ACVSYY325 pKa = 10.68GATGLVNNFKK335 pKa = 10.02IHH337 pKa = 6.34PNFYY341 pKa = 10.9LLLNSYY347 pKa = 10.31CRR349 pKa = 11.84INFKK353 pKa = 10.53PIVSALTRR361 pKa = 11.84SRR363 pKa = 11.84ISYY366 pKa = 9.71ISNKK370 pKa = 8.23MPVMPTEE377 pKa = 4.7DD378 pKa = 2.95MSAYY382 pKa = 9.73TKK384 pKa = 8.16MTMWLRR390 pKa = 11.84EE391 pKa = 3.54VDD393 pKa = 4.18RR394 pKa = 11.84IHH396 pKa = 7.09NGTEE400 pKa = 3.83SKK402 pKa = 10.58LRR404 pKa = 11.84NSSIIIIADD413 pKa = 3.27KK414 pKa = 11.16GSGKK418 pKa = 6.58TTCTKK423 pKa = 10.8YY424 pKa = 10.92LLQQLRR430 pKa = 11.84DD431 pKa = 3.69TSPSSKK437 pKa = 8.3TWGRR441 pKa = 11.84VDD443 pKa = 3.52SDD445 pKa = 5.29AFGKK449 pKa = 9.29WLSKK453 pKa = 9.74RR454 pKa = 11.84RR455 pKa = 11.84SMGMRR460 pKa = 11.84STDD463 pKa = 2.82ILLSNWQEE471 pKa = 3.7LDD473 pKa = 3.12ALQNDD478 pKa = 4.17EE479 pKa = 5.5SIPTYY484 pKa = 9.81FAHH487 pKa = 7.26EE488 pKa = 4.36MEE490 pKa = 4.74QMLLSAGMVEE500 pKa = 4.43YY501 pKa = 10.76GFDD504 pKa = 3.73PLSEE508 pKa = 4.08LRR510 pKa = 11.84ILDD513 pKa = 3.5EE514 pKa = 4.54FRR516 pKa = 11.84PIIYY520 pKa = 9.0TIINDD525 pKa = 3.8EE526 pKa = 4.41EE527 pKa = 3.95EE528 pKa = 4.29GLAAFYY534 pKa = 11.43NEE536 pKa = 4.0LDD538 pKa = 3.7KK539 pKa = 11.76FEE541 pKa = 5.47DD542 pKa = 3.94LPIGLMLEE550 pKa = 3.97SHH552 pKa = 6.79TDD554 pKa = 3.43VEE556 pKa = 4.23ISKK559 pKa = 9.3MFGTGHH565 pKa = 6.98IVTLDD570 pKa = 3.23APYY573 pKa = 10.33NAEE576 pKa = 3.75AAVADD581 pKa = 4.78RR582 pKa = 11.84KK583 pKa = 10.16RR584 pKa = 11.84SKK586 pKa = 10.5ISINVEE592 pKa = 3.64LMLFRR597 pKa = 11.84AWSRR601 pKa = 11.84LRR603 pKa = 11.84VSTYY607 pKa = 10.91PMLRR611 pKa = 11.84TQAFFDD617 pKa = 3.89TQLLAIASDD626 pKa = 3.67QEE628 pKa = 4.4TNPRR632 pKa = 3.56
Molecular weight: 72.76 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7971 |
329 |
1301 |
885.7 |
100.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.319 ± 0.367 | 1.104 ± 0.281 |
6.197 ± 0.24 | 6.122 ± 0.283 |
4.215 ± 0.187 | 5.018 ± 0.263 |
1.882 ± 0.122 | 7.916 ± 0.171 |
5.721 ± 0.248 | 8.744 ± 0.403 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.785 ± 0.241 | 5.545 ± 0.315 |
4.391 ± 0.433 | 4.303 ± 0.27 |
5.131 ± 0.216 | 7.377 ± 0.393 |
7.327 ± 0.297 | 5.633 ± 0.259 |
1.28 ± 0.161 | 3.989 ± 0.232 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
