
Ganoderma sinense ZZ0214-1
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetes incertae sedis; Polyporales; Polyporaceae; Ganoderma; Ganoderma sinense
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
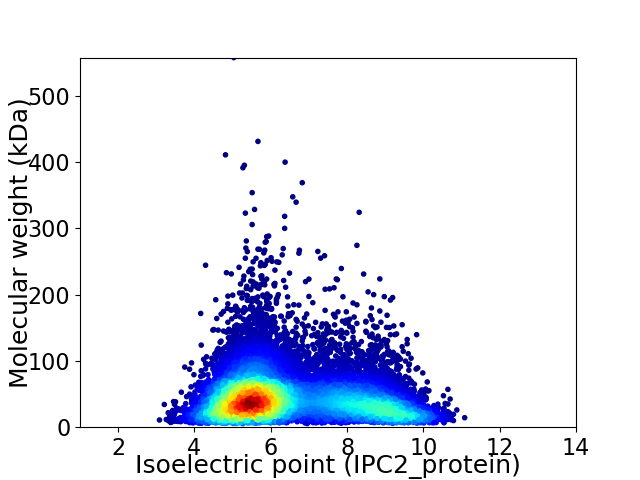
Virtual 2D-PAGE plot for 15394 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2G8S310|A0A2G8S310_9APHY Transcription factor OS=Ganoderma sinense ZZ0214-1 OX=1077348 GN=GSI_09696 PE=4 SV=1
MM1 pKa = 7.46FLPLTLLAALPLFPVWAAPMCSLKK25 pKa = 10.53PPKK28 pKa = 9.89SASATSSAPAPTGTTVGATGSNSNGAQVVATTWYY62 pKa = 10.16AGWHH66 pKa = 5.46GTDD69 pKa = 4.58FPPEE73 pKa = 3.64NVTWSKK79 pKa = 11.41YY80 pKa = 9.46SAATYY85 pKa = 10.39AFAVTTPDD93 pKa = 3.2VNTVSLADD101 pKa = 3.66SDD103 pKa = 5.11EE104 pKa = 4.12EE105 pKa = 4.08LLPRR109 pKa = 11.84FVDD112 pKa = 3.58QAHH115 pKa = 5.76QNDD118 pKa = 3.85VRR120 pKa = 11.84AMLTIGGWTGSQYY133 pKa = 10.5FSSAVATEE141 pKa = 4.03ANRR144 pKa = 11.84TAFVKK149 pKa = 10.14TMTNLISKK157 pKa = 10.05YY158 pKa = 10.5NLDD161 pKa = 3.82GLDD164 pKa = 4.4FDD166 pKa = 4.25WEE168 pKa = 4.49YY169 pKa = 10.89PNKK172 pKa = 10.11QGAGCNIISDD182 pKa = 3.79NDD184 pKa = 3.74SQNFLSFLQSLRR196 pKa = 11.84SAVGPDD202 pKa = 3.31VTLTAAAGITPFAGTDD218 pKa = 3.65GTPMTDD224 pKa = 2.45VSGFAKK230 pKa = 10.52VLDD233 pKa = 3.68WVAIMNYY240 pKa = 9.44DD241 pKa = 3.06VWGSWSSSVGPNAPLNDD258 pKa = 3.78TCASAADD265 pKa = 3.85QEE267 pKa = 4.83GSAVSAVKK275 pKa = 10.19AWTTAGFPANQLLLGVAAYY294 pKa = 9.26GHH296 pKa = 6.14SFSVPTTDD304 pKa = 4.86ALASSSSSNTDD315 pKa = 2.68SSQSALTAYY324 pKa = 9.45PAFDD328 pKa = 4.78KK329 pKa = 11.2SNQPMGDD336 pKa = 2.73SWDD339 pKa = 3.76AVSAEE344 pKa = 3.81SDD346 pKa = 3.26MCGVVTAAGPSGIFDD361 pKa = 3.43FWGLVEE367 pKa = 5.63GGFLTTNGTAASGIDD382 pKa = 3.6YY383 pKa = 10.8RR384 pKa = 11.84FDD386 pKa = 3.5ACSQTPYY393 pKa = 10.45VYY395 pKa = 10.71NPSTQVMVSYY405 pKa = 11.36DD406 pKa = 3.57DD407 pKa = 3.73TRR409 pKa = 11.84SFAAKK414 pKa = 10.07GKK416 pKa = 9.98FINDD420 pKa = 3.08QGLLGFAMWEE430 pKa = 3.78AAGDD434 pKa = 4.03YY435 pKa = 10.91KK436 pKa = 11.72DD437 pKa = 4.18MLLDD441 pKa = 5.26AISDD445 pKa = 4.07AIGIVEE451 pKa = 4.12VSCC454 pKa = 5.0
MM1 pKa = 7.46FLPLTLLAALPLFPVWAAPMCSLKK25 pKa = 10.53PPKK28 pKa = 9.89SASATSSAPAPTGTTVGATGSNSNGAQVVATTWYY62 pKa = 10.16AGWHH66 pKa = 5.46GTDD69 pKa = 4.58FPPEE73 pKa = 3.64NVTWSKK79 pKa = 11.41YY80 pKa = 9.46SAATYY85 pKa = 10.39AFAVTTPDD93 pKa = 3.2VNTVSLADD101 pKa = 3.66SDD103 pKa = 5.11EE104 pKa = 4.12EE105 pKa = 4.08LLPRR109 pKa = 11.84FVDD112 pKa = 3.58QAHH115 pKa = 5.76QNDD118 pKa = 3.85VRR120 pKa = 11.84AMLTIGGWTGSQYY133 pKa = 10.5FSSAVATEE141 pKa = 4.03ANRR144 pKa = 11.84TAFVKK149 pKa = 10.14TMTNLISKK157 pKa = 10.05YY158 pKa = 10.5NLDD161 pKa = 3.82GLDD164 pKa = 4.4FDD166 pKa = 4.25WEE168 pKa = 4.49YY169 pKa = 10.89PNKK172 pKa = 10.11QGAGCNIISDD182 pKa = 3.79NDD184 pKa = 3.74SQNFLSFLQSLRR196 pKa = 11.84SAVGPDD202 pKa = 3.31VTLTAAAGITPFAGTDD218 pKa = 3.65GTPMTDD224 pKa = 2.45VSGFAKK230 pKa = 10.52VLDD233 pKa = 3.68WVAIMNYY240 pKa = 9.44DD241 pKa = 3.06VWGSWSSSVGPNAPLNDD258 pKa = 3.78TCASAADD265 pKa = 3.85QEE267 pKa = 4.83GSAVSAVKK275 pKa = 10.19AWTTAGFPANQLLLGVAAYY294 pKa = 9.26GHH296 pKa = 6.14SFSVPTTDD304 pKa = 4.86ALASSSSSNTDD315 pKa = 2.68SSQSALTAYY324 pKa = 9.45PAFDD328 pKa = 4.78KK329 pKa = 11.2SNQPMGDD336 pKa = 2.73SWDD339 pKa = 3.76AVSAEE344 pKa = 3.81SDD346 pKa = 3.26MCGVVTAAGPSGIFDD361 pKa = 3.43FWGLVEE367 pKa = 5.63GGFLTTNGTAASGIDD382 pKa = 3.6YY383 pKa = 10.8RR384 pKa = 11.84FDD386 pKa = 3.5ACSQTPYY393 pKa = 10.45VYY395 pKa = 10.71NPSTQVMVSYY405 pKa = 11.36DD406 pKa = 3.57DD407 pKa = 3.73TRR409 pKa = 11.84SFAAKK414 pKa = 10.07GKK416 pKa = 9.98FINDD420 pKa = 3.08QGLLGFAMWEE430 pKa = 3.78AAGDD434 pKa = 4.03YY435 pKa = 10.91KK436 pKa = 11.72DD437 pKa = 4.18MLLDD441 pKa = 5.26AISDD445 pKa = 4.07AIGIVEE451 pKa = 4.12VSCC454 pKa = 5.0
Molecular weight: 47.61 kDa
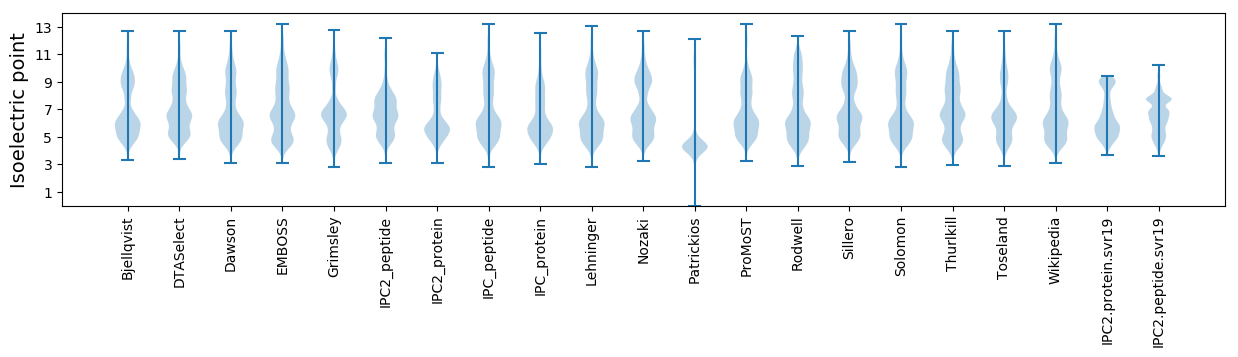
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2G8SQV3|A0A2G8SQV3_9APHY Uncharacterized protein OS=Ganoderma sinense ZZ0214-1 OX=1077348 GN=GSI_01811 PE=4 SV=1
MM1 pKa = 7.41PRR3 pKa = 11.84IPPSLARR10 pKa = 11.84LLSRR14 pKa = 11.84PPSLPAAVPRR24 pKa = 11.84PATSVSFASSHH35 pKa = 4.86TPFRR39 pKa = 11.84PILPSATPTSSLLRR53 pKa = 11.84LLPTLMTRR61 pKa = 11.84FNSVPSTLSPMFTPLLQVRR80 pKa = 11.84HH81 pKa = 4.5NTKK84 pKa = 9.7GQEE87 pKa = 4.08YY88 pKa = 9.97QPSQRR93 pKa = 11.84KK94 pKa = 8.85RR95 pKa = 11.84KK96 pKa = 9.01RR97 pKa = 11.84KK98 pKa = 9.64HH99 pKa = 5.8GFLTRR104 pKa = 11.84KK105 pKa = 9.7RR106 pKa = 11.84SVNGRR111 pKa = 11.84KK112 pKa = 8.41MLARR116 pKa = 11.84RR117 pKa = 11.84RR118 pKa = 11.84AKK120 pKa = 10.13GRR122 pKa = 11.84LFLSHH127 pKa = 7.07
MM1 pKa = 7.41PRR3 pKa = 11.84IPPSLARR10 pKa = 11.84LLSRR14 pKa = 11.84PPSLPAAVPRR24 pKa = 11.84PATSVSFASSHH35 pKa = 4.86TPFRR39 pKa = 11.84PILPSATPTSSLLRR53 pKa = 11.84LLPTLMTRR61 pKa = 11.84FNSVPSTLSPMFTPLLQVRR80 pKa = 11.84HH81 pKa = 4.5NTKK84 pKa = 9.7GQEE87 pKa = 4.08YY88 pKa = 9.97QPSQRR93 pKa = 11.84KK94 pKa = 8.85RR95 pKa = 11.84KK96 pKa = 9.01RR97 pKa = 11.84KK98 pKa = 9.64HH99 pKa = 5.8GFLTRR104 pKa = 11.84KK105 pKa = 9.7RR106 pKa = 11.84SVNGRR111 pKa = 11.84KK112 pKa = 8.41MLARR116 pKa = 11.84RR117 pKa = 11.84RR118 pKa = 11.84AKK120 pKa = 10.13GRR122 pKa = 11.84LFLSHH127 pKa = 7.07
Molecular weight: 14.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6730433 |
51 |
5028 |
437.2 |
48.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.393 ± 0.019 | 1.264 ± 0.007 |
5.752 ± 0.013 | 5.788 ± 0.018 |
3.678 ± 0.011 | 6.681 ± 0.017 |
2.637 ± 0.009 | 4.265 ± 0.013 |
3.994 ± 0.017 | 9.251 ± 0.023 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.012 ± 0.007 | 3.02 ± 0.01 |
7.061 ± 0.028 | 3.52 ± 0.012 |
6.601 ± 0.018 | 8.396 ± 0.025 |
5.977 ± 0.013 | 6.639 ± 0.014 |
1.487 ± 0.007 | 2.584 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
