
Beihai shrimp virus 3
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.81
Get precalculated fractions of proteins
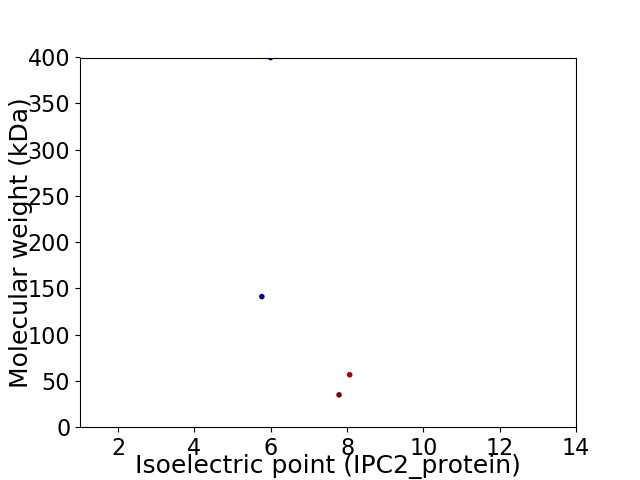
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KPD6|A0A1L3KPD6_9VIRU Uncharacterized protein OS=Beihai shrimp virus 3 OX=1922669 PE=4 SV=1
MM1 pKa = 7.9AEE3 pKa = 3.9TFLAILLLLLWCSPLPVLAKK23 pKa = 10.28GYY25 pKa = 9.14SPHH28 pKa = 5.11VTADD32 pKa = 3.07KK33 pKa = 9.66EE34 pKa = 4.38RR35 pKa = 11.84NMVRR39 pKa = 11.84VGAFTPRR46 pKa = 11.84SEE48 pKa = 4.37CTVTIKK54 pKa = 10.78PSGPCAWTADD64 pKa = 3.89GVGHH68 pKa = 6.95FDD70 pKa = 3.54GKK72 pKa = 10.21NVSWCPAYY80 pKa = 9.77PGNCDD85 pKa = 3.07LRR87 pKa = 11.84KK88 pKa = 7.15YY89 pKa = 9.59TEE91 pKa = 4.12MNGMRR96 pKa = 11.84TWSGCKK102 pKa = 9.56RR103 pKa = 11.84YY104 pKa = 9.99AVGNKK109 pKa = 9.36VGMCDD114 pKa = 3.78VEE116 pKa = 5.74GEE118 pKa = 4.58TFDD121 pKa = 5.18HH122 pKa = 6.63MEE124 pKa = 4.41GVCKK128 pKa = 10.41SVTEE132 pKa = 4.23MEE134 pKa = 4.62NDD136 pKa = 3.31PCLLFWGDD144 pKa = 3.25GTANEE149 pKa = 4.15ILAATEE155 pKa = 3.88QLEE158 pKa = 4.3PIYY161 pKa = 10.57TGSNYY166 pKa = 10.67KK167 pKa = 9.7MFVIEE172 pKa = 4.05WKK174 pKa = 9.03KK175 pKa = 10.78VKK177 pKa = 8.92WQFHH181 pKa = 5.86ISDD184 pKa = 3.95GGSLFRR190 pKa = 11.84MCRR193 pKa = 11.84TAWDD197 pKa = 4.3HH198 pKa = 7.15SCGTPCCRR206 pKa = 11.84AVPEE210 pKa = 4.42GEE212 pKa = 4.21RR213 pKa = 11.84LVCLITDD220 pKa = 4.02DD221 pKa = 5.81LYY223 pKa = 11.43EE224 pKa = 4.28KK225 pKa = 10.33CQYY228 pKa = 10.41QIRR231 pKa = 11.84QEE233 pKa = 4.0DD234 pKa = 4.13EE235 pKa = 3.57KK236 pKa = 11.03LQRR239 pKa = 11.84YY240 pKa = 8.72PEE242 pKa = 4.75DD243 pKa = 3.67LDD245 pKa = 3.46KK246 pKa = 11.35TEE248 pKa = 4.25PSGYY252 pKa = 10.53LPVDD256 pKa = 4.6LDD258 pKa = 3.94TKK260 pKa = 10.81DD261 pKa = 3.95LQHH264 pKa = 6.75HH265 pKa = 5.82SEE267 pKa = 4.15VHH269 pKa = 5.68FSEE272 pKa = 5.25LDD274 pKa = 3.45EE275 pKa = 5.19LVDD278 pKa = 5.6SEE280 pKa = 5.13ISCQRR285 pKa = 11.84PPLICGSRR293 pKa = 11.84RR294 pKa = 11.84YY295 pKa = 8.22TDD297 pKa = 3.3YY298 pKa = 10.94KK299 pKa = 9.69VSEE302 pKa = 4.37RR303 pKa = 11.84EE304 pKa = 3.5VWAPSTPADD313 pKa = 3.92SPNGAFKK320 pKa = 9.81CTPTDD325 pKa = 3.56PQTTSVEE332 pKa = 4.16CHH334 pKa = 5.57NFCLVKK340 pKa = 10.77ASGLCSGSEE349 pKa = 4.05SFGKK353 pKa = 10.05FFKK356 pKa = 10.53IGGGFSCHH364 pKa = 6.89CDD366 pKa = 2.65WVTKK370 pKa = 10.53ISTEE374 pKa = 4.03TIYY377 pKa = 10.74TVGTSIYY384 pKa = 9.68YY385 pKa = 10.31GEE387 pKa = 4.66HH388 pKa = 6.44EE389 pKa = 4.68CTVGAACAAANNTMRR404 pKa = 11.84RR405 pKa = 11.84IGRR408 pKa = 11.84RR409 pKa = 11.84LHH411 pKa = 6.6GVTEE415 pKa = 3.98EE416 pKa = 4.49LKK418 pKa = 11.0CKK420 pKa = 9.9STDD423 pKa = 3.37GQVDD427 pKa = 3.73CGIAEE432 pKa = 4.41CMFLVSAGNSSAVGRR447 pKa = 11.84AGDD450 pKa = 3.79QIKK453 pKa = 10.78LSVGLRR459 pKa = 11.84DD460 pKa = 3.7RR461 pKa = 11.84LRR463 pKa = 11.84GYY465 pKa = 9.85YY466 pKa = 10.17LQVKK470 pKa = 7.62LLCDD474 pKa = 3.83TPVMAGLKK482 pKa = 7.95TLKK485 pKa = 10.19KK486 pKa = 10.1QRR488 pKa = 11.84NEE490 pKa = 3.55NHH492 pKa = 5.37GSYY495 pKa = 10.67RR496 pKa = 11.84LFVPPEE502 pKa = 4.03EE503 pKa = 5.08LCDD506 pKa = 3.93CLGLRR511 pKa = 11.84SLLLDD516 pKa = 3.56DD517 pKa = 4.85CRR519 pKa = 11.84SLRR522 pKa = 11.84SAIVAMGTSAVVEE535 pKa = 4.56FLAILWCIVLIVWILSVVKK554 pKa = 10.51KK555 pKa = 9.86FLYY558 pKa = 10.25RR559 pKa = 11.84AGFLLDD565 pKa = 3.51GRR567 pKa = 11.84SVSNEE572 pKa = 3.45LLEE575 pKa = 4.32CRR577 pKa = 11.84EE578 pKa = 4.1TGEE581 pKa = 5.06LRR583 pKa = 11.84DD584 pKa = 3.64EE585 pKa = 4.01QVRR588 pKa = 11.84IIKK591 pKa = 10.34LYY593 pKa = 10.88FEE595 pKa = 4.9DD596 pKa = 6.23AITKK600 pKa = 10.4HH601 pKa = 5.63MFCSLGWWTRR611 pKa = 11.84RR612 pKa = 11.84QQRR615 pKa = 11.84ALKK618 pKa = 9.42KK619 pKa = 9.95VCLSYY624 pKa = 11.15YY625 pKa = 10.21LYY627 pKa = 10.9LIIEE631 pKa = 5.17GYY633 pKa = 9.86GKK635 pKa = 10.18NDD637 pKa = 2.27AWQRR641 pKa = 11.84SCKK644 pKa = 9.25IWKK647 pKa = 8.65QAAAGNEE654 pKa = 4.19LKK656 pKa = 11.11DD657 pKa = 3.91EE658 pKa = 4.17VLKK661 pKa = 11.24DD662 pKa = 3.68LVTSGSRR669 pKa = 11.84KK670 pKa = 8.95IGARR674 pKa = 11.84SDD676 pKa = 3.51QKK678 pKa = 9.89TLNPASLSGVLVTLVYY694 pKa = 10.77CFTLMRR700 pKa = 11.84LVGGSCEE707 pKa = 4.22TGSVDD712 pKa = 4.15VSSSVCTMASEE723 pKa = 4.53DD724 pKa = 3.86TMSCSVKK731 pKa = 8.32TTILSTIDD739 pKa = 3.31VVGKK743 pKa = 7.39STCFNLQNKK752 pKa = 5.87TTGDD756 pKa = 2.79IVGQVEE762 pKa = 4.46VEE764 pKa = 3.85YY765 pKa = 11.69SMLEE769 pKa = 4.22RR770 pKa = 11.84AWSLNNLYY778 pKa = 8.69YY779 pKa = 10.05TGHH782 pKa = 6.68WSLMTRR788 pKa = 11.84EE789 pKa = 4.97DD790 pKa = 3.53SSCYY794 pKa = 9.64NKK796 pKa = 10.2GWCTSKK802 pKa = 10.85CDD804 pKa = 4.38GKK806 pKa = 10.33MKK808 pKa = 9.98PAGYY812 pKa = 10.18RR813 pKa = 11.84NQLCVNGICKK823 pKa = 8.67VQPVSSACCYY833 pKa = 9.33WYY835 pKa = 9.18VTIFTCGSSYY845 pKa = 11.1KK846 pKa = 10.32CGGVTSMIRR855 pKa = 11.84TEE857 pKa = 3.87TDD859 pKa = 3.23GNHH862 pKa = 5.11GWVKK866 pKa = 10.38GVGRR870 pKa = 11.84GEE872 pKa = 4.29ALTTVAVSVRR882 pKa = 11.84TNGGDD887 pKa = 3.44LKK889 pKa = 11.11EE890 pKa = 4.29SDD892 pKa = 3.96GVIKK896 pKa = 10.75GVGTHH901 pKa = 6.36KK902 pKa = 10.6SGPITVRR909 pKa = 11.84VKK911 pKa = 10.78GVANLPQINFGGHH924 pKa = 6.86KK925 pKa = 9.75IYY927 pKa = 9.98ATDD930 pKa = 3.25TATYY934 pKa = 7.93LTLASDD940 pKa = 3.73INNPVVGTIGDD951 pKa = 3.91VQASSAGAWTNPPEE965 pKa = 4.9AITGEE970 pKa = 3.91EE971 pKa = 4.01HH972 pKa = 7.27LEE974 pKa = 4.0HH975 pKa = 6.84FRR977 pKa = 11.84TADD980 pKa = 3.66VVDD983 pKa = 5.51LSCSSWDD990 pKa = 3.44MSILQSEE997 pKa = 4.24PGINLLHH1004 pKa = 6.75DD1005 pKa = 3.83QFSDD1009 pKa = 3.37TQLPVVFQGSAFKK1022 pKa = 10.47EE1023 pKa = 4.28VSNEE1027 pKa = 3.65RR1028 pKa = 11.84LIEE1031 pKa = 4.13EE1032 pKa = 4.46LTGSRR1037 pKa = 11.84PLSVEE1042 pKa = 3.52IAISGTFAVKK1052 pKa = 10.15HH1053 pKa = 5.54VIKK1056 pKa = 10.11EE1057 pKa = 4.37CKK1059 pKa = 9.5PEE1061 pKa = 3.86MEE1063 pKa = 4.2FVEE1066 pKa = 4.43MKK1068 pKa = 10.82GCFSCYY1074 pKa = 10.01NPGAILTVRR1083 pKa = 11.84TRR1085 pKa = 11.84SNNPYY1090 pKa = 9.94SCVAALTTNSEE1101 pKa = 4.35HH1102 pKa = 6.35FNCPGSLEE1110 pKa = 4.15FEE1112 pKa = 4.8PGDD1115 pKa = 3.48FVEE1118 pKa = 5.55RR1119 pKa = 11.84KK1120 pKa = 8.87VSCISSEE1127 pKa = 4.12EE1128 pKa = 4.05EE1129 pKa = 3.42NDD1131 pKa = 3.52IVLTAQTRR1139 pKa = 11.84TKK1141 pKa = 10.8VSVEE1145 pKa = 3.85VTGTLLADD1153 pKa = 3.92RR1154 pKa = 11.84LLAANEE1160 pKa = 4.16LEE1162 pKa = 4.22GAEE1165 pKa = 4.26PDD1167 pKa = 3.59EE1168 pKa = 4.93SSSVLNPLGLSDD1180 pKa = 5.83LNITLISIGSVTTFVVIFILLACFCGPFRR1209 pKa = 11.84DD1210 pKa = 4.17CLVFACPCLRR1220 pKa = 11.84SLSRR1224 pKa = 11.84KK1225 pKa = 9.99LPDD1228 pKa = 3.58KK1229 pKa = 11.24LSGFKK1234 pKa = 10.79DD1235 pKa = 3.44KK1236 pKa = 10.56IDD1238 pKa = 3.79RR1239 pKa = 11.84SAEE1242 pKa = 3.86KK1243 pKa = 10.66AKK1245 pKa = 10.97VSGIKK1250 pKa = 10.24AKK1252 pKa = 9.96SEE1254 pKa = 4.21KK1255 pKa = 10.15EE1256 pKa = 3.36WGEE1259 pKa = 3.79AKK1261 pKa = 10.26KK1262 pKa = 10.71AQDD1265 pKa = 3.27NHH1267 pKa = 6.37DD1268 pKa = 3.49KK1269 pKa = 10.19VVRR1272 pKa = 11.84MRR1274 pKa = 11.84KK1275 pKa = 9.6KK1276 pKa = 10.44FGQAA1280 pKa = 2.79
MM1 pKa = 7.9AEE3 pKa = 3.9TFLAILLLLLWCSPLPVLAKK23 pKa = 10.28GYY25 pKa = 9.14SPHH28 pKa = 5.11VTADD32 pKa = 3.07KK33 pKa = 9.66EE34 pKa = 4.38RR35 pKa = 11.84NMVRR39 pKa = 11.84VGAFTPRR46 pKa = 11.84SEE48 pKa = 4.37CTVTIKK54 pKa = 10.78PSGPCAWTADD64 pKa = 3.89GVGHH68 pKa = 6.95FDD70 pKa = 3.54GKK72 pKa = 10.21NVSWCPAYY80 pKa = 9.77PGNCDD85 pKa = 3.07LRR87 pKa = 11.84KK88 pKa = 7.15YY89 pKa = 9.59TEE91 pKa = 4.12MNGMRR96 pKa = 11.84TWSGCKK102 pKa = 9.56RR103 pKa = 11.84YY104 pKa = 9.99AVGNKK109 pKa = 9.36VGMCDD114 pKa = 3.78VEE116 pKa = 5.74GEE118 pKa = 4.58TFDD121 pKa = 5.18HH122 pKa = 6.63MEE124 pKa = 4.41GVCKK128 pKa = 10.41SVTEE132 pKa = 4.23MEE134 pKa = 4.62NDD136 pKa = 3.31PCLLFWGDD144 pKa = 3.25GTANEE149 pKa = 4.15ILAATEE155 pKa = 3.88QLEE158 pKa = 4.3PIYY161 pKa = 10.57TGSNYY166 pKa = 10.67KK167 pKa = 9.7MFVIEE172 pKa = 4.05WKK174 pKa = 9.03KK175 pKa = 10.78VKK177 pKa = 8.92WQFHH181 pKa = 5.86ISDD184 pKa = 3.95GGSLFRR190 pKa = 11.84MCRR193 pKa = 11.84TAWDD197 pKa = 4.3HH198 pKa = 7.15SCGTPCCRR206 pKa = 11.84AVPEE210 pKa = 4.42GEE212 pKa = 4.21RR213 pKa = 11.84LVCLITDD220 pKa = 4.02DD221 pKa = 5.81LYY223 pKa = 11.43EE224 pKa = 4.28KK225 pKa = 10.33CQYY228 pKa = 10.41QIRR231 pKa = 11.84QEE233 pKa = 4.0DD234 pKa = 4.13EE235 pKa = 3.57KK236 pKa = 11.03LQRR239 pKa = 11.84YY240 pKa = 8.72PEE242 pKa = 4.75DD243 pKa = 3.67LDD245 pKa = 3.46KK246 pKa = 11.35TEE248 pKa = 4.25PSGYY252 pKa = 10.53LPVDD256 pKa = 4.6LDD258 pKa = 3.94TKK260 pKa = 10.81DD261 pKa = 3.95LQHH264 pKa = 6.75HH265 pKa = 5.82SEE267 pKa = 4.15VHH269 pKa = 5.68FSEE272 pKa = 5.25LDD274 pKa = 3.45EE275 pKa = 5.19LVDD278 pKa = 5.6SEE280 pKa = 5.13ISCQRR285 pKa = 11.84PPLICGSRR293 pKa = 11.84RR294 pKa = 11.84YY295 pKa = 8.22TDD297 pKa = 3.3YY298 pKa = 10.94KK299 pKa = 9.69VSEE302 pKa = 4.37RR303 pKa = 11.84EE304 pKa = 3.5VWAPSTPADD313 pKa = 3.92SPNGAFKK320 pKa = 9.81CTPTDD325 pKa = 3.56PQTTSVEE332 pKa = 4.16CHH334 pKa = 5.57NFCLVKK340 pKa = 10.77ASGLCSGSEE349 pKa = 4.05SFGKK353 pKa = 10.05FFKK356 pKa = 10.53IGGGFSCHH364 pKa = 6.89CDD366 pKa = 2.65WVTKK370 pKa = 10.53ISTEE374 pKa = 4.03TIYY377 pKa = 10.74TVGTSIYY384 pKa = 9.68YY385 pKa = 10.31GEE387 pKa = 4.66HH388 pKa = 6.44EE389 pKa = 4.68CTVGAACAAANNTMRR404 pKa = 11.84RR405 pKa = 11.84IGRR408 pKa = 11.84RR409 pKa = 11.84LHH411 pKa = 6.6GVTEE415 pKa = 3.98EE416 pKa = 4.49LKK418 pKa = 11.0CKK420 pKa = 9.9STDD423 pKa = 3.37GQVDD427 pKa = 3.73CGIAEE432 pKa = 4.41CMFLVSAGNSSAVGRR447 pKa = 11.84AGDD450 pKa = 3.79QIKK453 pKa = 10.78LSVGLRR459 pKa = 11.84DD460 pKa = 3.7RR461 pKa = 11.84LRR463 pKa = 11.84GYY465 pKa = 9.85YY466 pKa = 10.17LQVKK470 pKa = 7.62LLCDD474 pKa = 3.83TPVMAGLKK482 pKa = 7.95TLKK485 pKa = 10.19KK486 pKa = 10.1QRR488 pKa = 11.84NEE490 pKa = 3.55NHH492 pKa = 5.37GSYY495 pKa = 10.67RR496 pKa = 11.84LFVPPEE502 pKa = 4.03EE503 pKa = 5.08LCDD506 pKa = 3.93CLGLRR511 pKa = 11.84SLLLDD516 pKa = 3.56DD517 pKa = 4.85CRR519 pKa = 11.84SLRR522 pKa = 11.84SAIVAMGTSAVVEE535 pKa = 4.56FLAILWCIVLIVWILSVVKK554 pKa = 10.51KK555 pKa = 9.86FLYY558 pKa = 10.25RR559 pKa = 11.84AGFLLDD565 pKa = 3.51GRR567 pKa = 11.84SVSNEE572 pKa = 3.45LLEE575 pKa = 4.32CRR577 pKa = 11.84EE578 pKa = 4.1TGEE581 pKa = 5.06LRR583 pKa = 11.84DD584 pKa = 3.64EE585 pKa = 4.01QVRR588 pKa = 11.84IIKK591 pKa = 10.34LYY593 pKa = 10.88FEE595 pKa = 4.9DD596 pKa = 6.23AITKK600 pKa = 10.4HH601 pKa = 5.63MFCSLGWWTRR611 pKa = 11.84RR612 pKa = 11.84QQRR615 pKa = 11.84ALKK618 pKa = 9.42KK619 pKa = 9.95VCLSYY624 pKa = 11.15YY625 pKa = 10.21LYY627 pKa = 10.9LIIEE631 pKa = 5.17GYY633 pKa = 9.86GKK635 pKa = 10.18NDD637 pKa = 2.27AWQRR641 pKa = 11.84SCKK644 pKa = 9.25IWKK647 pKa = 8.65QAAAGNEE654 pKa = 4.19LKK656 pKa = 11.11DD657 pKa = 3.91EE658 pKa = 4.17VLKK661 pKa = 11.24DD662 pKa = 3.68LVTSGSRR669 pKa = 11.84KK670 pKa = 8.95IGARR674 pKa = 11.84SDD676 pKa = 3.51QKK678 pKa = 9.89TLNPASLSGVLVTLVYY694 pKa = 10.77CFTLMRR700 pKa = 11.84LVGGSCEE707 pKa = 4.22TGSVDD712 pKa = 4.15VSSSVCTMASEE723 pKa = 4.53DD724 pKa = 3.86TMSCSVKK731 pKa = 8.32TTILSTIDD739 pKa = 3.31VVGKK743 pKa = 7.39STCFNLQNKK752 pKa = 5.87TTGDD756 pKa = 2.79IVGQVEE762 pKa = 4.46VEE764 pKa = 3.85YY765 pKa = 11.69SMLEE769 pKa = 4.22RR770 pKa = 11.84AWSLNNLYY778 pKa = 8.69YY779 pKa = 10.05TGHH782 pKa = 6.68WSLMTRR788 pKa = 11.84EE789 pKa = 4.97DD790 pKa = 3.53SSCYY794 pKa = 9.64NKK796 pKa = 10.2GWCTSKK802 pKa = 10.85CDD804 pKa = 4.38GKK806 pKa = 10.33MKK808 pKa = 9.98PAGYY812 pKa = 10.18RR813 pKa = 11.84NQLCVNGICKK823 pKa = 8.67VQPVSSACCYY833 pKa = 9.33WYY835 pKa = 9.18VTIFTCGSSYY845 pKa = 11.1KK846 pKa = 10.32CGGVTSMIRR855 pKa = 11.84TEE857 pKa = 3.87TDD859 pKa = 3.23GNHH862 pKa = 5.11GWVKK866 pKa = 10.38GVGRR870 pKa = 11.84GEE872 pKa = 4.29ALTTVAVSVRR882 pKa = 11.84TNGGDD887 pKa = 3.44LKK889 pKa = 11.11EE890 pKa = 4.29SDD892 pKa = 3.96GVIKK896 pKa = 10.75GVGTHH901 pKa = 6.36KK902 pKa = 10.6SGPITVRR909 pKa = 11.84VKK911 pKa = 10.78GVANLPQINFGGHH924 pKa = 6.86KK925 pKa = 9.75IYY927 pKa = 9.98ATDD930 pKa = 3.25TATYY934 pKa = 7.93LTLASDD940 pKa = 3.73INNPVVGTIGDD951 pKa = 3.91VQASSAGAWTNPPEE965 pKa = 4.9AITGEE970 pKa = 3.91EE971 pKa = 4.01HH972 pKa = 7.27LEE974 pKa = 4.0HH975 pKa = 6.84FRR977 pKa = 11.84TADD980 pKa = 3.66VVDD983 pKa = 5.51LSCSSWDD990 pKa = 3.44MSILQSEE997 pKa = 4.24PGINLLHH1004 pKa = 6.75DD1005 pKa = 3.83QFSDD1009 pKa = 3.37TQLPVVFQGSAFKK1022 pKa = 10.47EE1023 pKa = 4.28VSNEE1027 pKa = 3.65RR1028 pKa = 11.84LIEE1031 pKa = 4.13EE1032 pKa = 4.46LTGSRR1037 pKa = 11.84PLSVEE1042 pKa = 3.52IAISGTFAVKK1052 pKa = 10.15HH1053 pKa = 5.54VIKK1056 pKa = 10.11EE1057 pKa = 4.37CKK1059 pKa = 9.5PEE1061 pKa = 3.86MEE1063 pKa = 4.2FVEE1066 pKa = 4.43MKK1068 pKa = 10.82GCFSCYY1074 pKa = 10.01NPGAILTVRR1083 pKa = 11.84TRR1085 pKa = 11.84SNNPYY1090 pKa = 9.94SCVAALTTNSEE1101 pKa = 4.35HH1102 pKa = 6.35FNCPGSLEE1110 pKa = 4.15FEE1112 pKa = 4.8PGDD1115 pKa = 3.48FVEE1118 pKa = 5.55RR1119 pKa = 11.84KK1120 pKa = 8.87VSCISSEE1127 pKa = 4.12EE1128 pKa = 4.05EE1129 pKa = 3.42NDD1131 pKa = 3.52IVLTAQTRR1139 pKa = 11.84TKK1141 pKa = 10.8VSVEE1145 pKa = 3.85VTGTLLADD1153 pKa = 3.92RR1154 pKa = 11.84LLAANEE1160 pKa = 4.16LEE1162 pKa = 4.22GAEE1165 pKa = 4.26PDD1167 pKa = 3.59EE1168 pKa = 4.93SSSVLNPLGLSDD1180 pKa = 5.83LNITLISIGSVTTFVVIFILLACFCGPFRR1209 pKa = 11.84DD1210 pKa = 4.17CLVFACPCLRR1220 pKa = 11.84SLSRR1224 pKa = 11.84KK1225 pKa = 9.99LPDD1228 pKa = 3.58KK1229 pKa = 11.24LSGFKK1234 pKa = 10.79DD1235 pKa = 3.44KK1236 pKa = 10.56IDD1238 pKa = 3.79RR1239 pKa = 11.84SAEE1242 pKa = 3.86KK1243 pKa = 10.66AKK1245 pKa = 10.97VSGIKK1250 pKa = 10.24AKK1252 pKa = 9.96SEE1254 pKa = 4.21KK1255 pKa = 10.15EE1256 pKa = 3.36WGEE1259 pKa = 3.79AKK1261 pKa = 10.26KK1262 pKa = 10.71AQDD1265 pKa = 3.27NHH1267 pKa = 6.37DD1268 pKa = 3.49KK1269 pKa = 10.19VVRR1272 pKa = 11.84MRR1274 pKa = 11.84KK1275 pKa = 9.6KK1276 pKa = 10.44FGQAA1280 pKa = 2.79
Molecular weight: 141.15 kDa
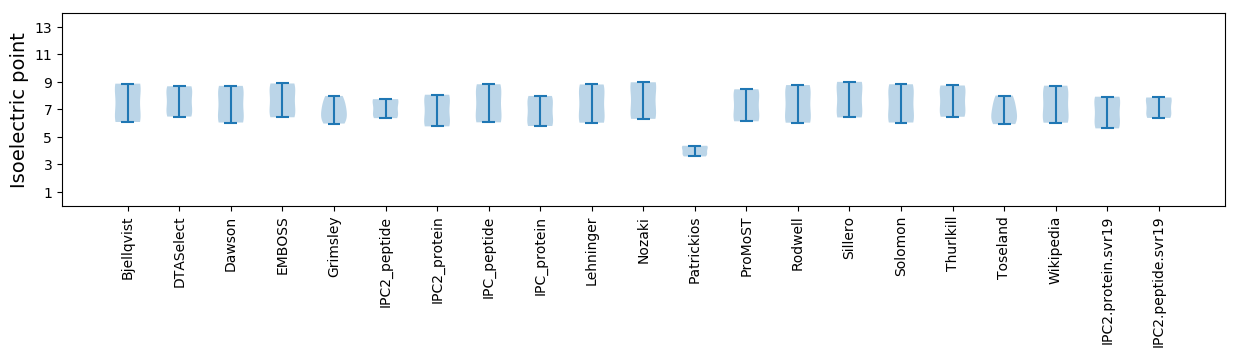
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KPD6|A0A1L3KPD6_9VIRU Uncharacterized protein OS=Beihai shrimp virus 3 OX=1922669 PE=4 SV=1
MM1 pKa = 7.69SSRR4 pKa = 11.84ISIKK8 pKa = 10.25YY9 pKa = 9.49LPEE12 pKa = 3.9VTFKK16 pKa = 10.97AANGEE21 pKa = 4.23ALFSPKK27 pKa = 9.9FEE29 pKa = 4.23RR30 pKa = 11.84TGVTSRR36 pKa = 11.84EE37 pKa = 4.25SLSGAVNRR45 pKa = 11.84ALNCMVRR52 pKa = 11.84VKK54 pKa = 10.61DD55 pKa = 3.94FQVVGHH61 pKa = 6.66AFITKK66 pKa = 10.12LIAAGHH72 pKa = 6.62DD73 pKa = 3.75PYY75 pKa = 11.72VDD77 pKa = 3.35AVVEE81 pKa = 3.99ALIQGDD87 pKa = 4.41DD88 pKa = 4.27DD89 pKa = 3.86NTLTTKK95 pKa = 11.04YY96 pKa = 8.9MVAMDD101 pKa = 4.27KK102 pKa = 10.99VAILSGIPKK111 pKa = 8.41EE112 pKa = 4.4TGSVATEE119 pKa = 3.94LKK121 pKa = 10.54VAASQLNSVNDD132 pKa = 4.93LISAMKK138 pKa = 9.9QNSSAGMSSTHH149 pKa = 5.81VVDD152 pKa = 4.95VYY154 pKa = 11.48TNVLNLSACCNGSTHH169 pKa = 7.16SYY171 pKa = 10.91LPDD174 pKa = 3.8LATNSKK180 pKa = 7.43GQKK183 pKa = 9.15MIVTAMVSRR192 pKa = 11.84PRR194 pKa = 11.84LNQRR198 pKa = 11.84DD199 pKa = 4.16RR200 pKa = 11.84DD201 pKa = 4.01LLDD204 pKa = 3.8AALNLIGLNSPDD216 pKa = 4.7HH217 pKa = 5.88IALYY221 pKa = 11.22NEE223 pKa = 4.26MQQMAGSGAAVVANNSKK240 pKa = 10.36NICYY244 pKa = 9.46PFYY247 pKa = 10.45ISLVAAMMFRR257 pKa = 11.84DD258 pKa = 3.96NKK260 pKa = 10.2TDD262 pKa = 3.26TDD264 pKa = 3.93VQSFLIPFVKK274 pKa = 9.51MAKK277 pKa = 8.11TKK279 pKa = 10.48RR280 pKa = 11.84GAATKK285 pKa = 10.21SHH287 pKa = 7.06NILQAYY293 pKa = 7.39NFPGYY298 pKa = 9.74IQGGKK303 pKa = 9.83DD304 pKa = 2.77IHH306 pKa = 7.15LNPCYY311 pKa = 9.16LTPYY315 pKa = 9.68RR316 pKa = 11.84MCRR319 pKa = 11.84ALMAAAPMPLVTNARR334 pKa = 11.84GIMMTPEE341 pKa = 4.33SIAFVLRR348 pKa = 11.84LYY350 pKa = 10.71EE351 pKa = 4.75IDD353 pKa = 3.83VEE355 pKa = 4.71LGGDD359 pKa = 3.66RR360 pKa = 11.84NINDD364 pKa = 3.4ATSIFHH370 pKa = 6.57HH371 pKa = 6.29MLVGGIGSYY380 pKa = 9.29ATTHH384 pKa = 6.04NKK386 pKa = 8.28TGNALNVTVKK396 pKa = 10.64LSGQNYY402 pKa = 7.87TPAPPMTQVMTVVAQPQPQRR422 pKa = 11.84AAAPMAPTAPPTAAVAPPPAPAPATDD448 pKa = 3.29RR449 pKa = 11.84LGNIVPTAPAPVPGTVPAPPAIAPLPGAIATSATTTVTVPQQQQPQQQQAPAQAPATTAAATPTVAVPGATAGISTNTATPSNAGNNSNAA539 pKa = 3.65
MM1 pKa = 7.69SSRR4 pKa = 11.84ISIKK8 pKa = 10.25YY9 pKa = 9.49LPEE12 pKa = 3.9VTFKK16 pKa = 10.97AANGEE21 pKa = 4.23ALFSPKK27 pKa = 9.9FEE29 pKa = 4.23RR30 pKa = 11.84TGVTSRR36 pKa = 11.84EE37 pKa = 4.25SLSGAVNRR45 pKa = 11.84ALNCMVRR52 pKa = 11.84VKK54 pKa = 10.61DD55 pKa = 3.94FQVVGHH61 pKa = 6.66AFITKK66 pKa = 10.12LIAAGHH72 pKa = 6.62DD73 pKa = 3.75PYY75 pKa = 11.72VDD77 pKa = 3.35AVVEE81 pKa = 3.99ALIQGDD87 pKa = 4.41DD88 pKa = 4.27DD89 pKa = 3.86NTLTTKK95 pKa = 11.04YY96 pKa = 8.9MVAMDD101 pKa = 4.27KK102 pKa = 10.99VAILSGIPKK111 pKa = 8.41EE112 pKa = 4.4TGSVATEE119 pKa = 3.94LKK121 pKa = 10.54VAASQLNSVNDD132 pKa = 4.93LISAMKK138 pKa = 9.9QNSSAGMSSTHH149 pKa = 5.81VVDD152 pKa = 4.95VYY154 pKa = 11.48TNVLNLSACCNGSTHH169 pKa = 7.16SYY171 pKa = 10.91LPDD174 pKa = 3.8LATNSKK180 pKa = 7.43GQKK183 pKa = 9.15MIVTAMVSRR192 pKa = 11.84PRR194 pKa = 11.84LNQRR198 pKa = 11.84DD199 pKa = 4.16RR200 pKa = 11.84DD201 pKa = 4.01LLDD204 pKa = 3.8AALNLIGLNSPDD216 pKa = 4.7HH217 pKa = 5.88IALYY221 pKa = 11.22NEE223 pKa = 4.26MQQMAGSGAAVVANNSKK240 pKa = 10.36NICYY244 pKa = 9.46PFYY247 pKa = 10.45ISLVAAMMFRR257 pKa = 11.84DD258 pKa = 3.96NKK260 pKa = 10.2TDD262 pKa = 3.26TDD264 pKa = 3.93VQSFLIPFVKK274 pKa = 9.51MAKK277 pKa = 8.11TKK279 pKa = 10.48RR280 pKa = 11.84GAATKK285 pKa = 10.21SHH287 pKa = 7.06NILQAYY293 pKa = 7.39NFPGYY298 pKa = 9.74IQGGKK303 pKa = 9.83DD304 pKa = 2.77IHH306 pKa = 7.15LNPCYY311 pKa = 9.16LTPYY315 pKa = 9.68RR316 pKa = 11.84MCRR319 pKa = 11.84ALMAAAPMPLVTNARR334 pKa = 11.84GIMMTPEE341 pKa = 4.33SIAFVLRR348 pKa = 11.84LYY350 pKa = 10.71EE351 pKa = 4.75IDD353 pKa = 3.83VEE355 pKa = 4.71LGGDD359 pKa = 3.66RR360 pKa = 11.84NINDD364 pKa = 3.4ATSIFHH370 pKa = 6.57HH371 pKa = 6.29MLVGGIGSYY380 pKa = 9.29ATTHH384 pKa = 6.04NKK386 pKa = 8.28TGNALNVTVKK396 pKa = 10.64LSGQNYY402 pKa = 7.87TPAPPMTQVMTVVAQPQPQRR422 pKa = 11.84AAAPMAPTAPPTAAVAPPPAPAPATDD448 pKa = 3.29RR449 pKa = 11.84LGNIVPTAPAPVPGTVPAPPAIAPLPGAIATSATTTVTVPQQQQPQQQQAPAQAPATTAAATPTVAVPGATAGISTNTATPSNAGNNSNAA539 pKa = 3.65
Molecular weight: 56.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5671 |
312 |
3540 |
1417.8 |
158.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.018 ± 1.056 | 2.451 ± 0.866 |
5.414 ± 0.47 | 6.648 ± 0.977 |
3.527 ± 0.242 | 5.678 ± 0.908 |
2.028 ± 0.075 | 4.761 ± 0.083 |
6.048 ± 0.362 | 9.646 ± 0.869 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.839 ± 0.305 | 4.109 ± 0.378 |
4.303 ± 0.783 | 2.998 ± 0.258 |
5.537 ± 0.716 | 8.852 ± 0.431 |
6.524 ± 0.393 | 7.371 ± 0.523 |
1.111 ± 0.279 | 3.139 ± 0.277 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
