
Candidatus Cytomitobacter primus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Holosporales; Holosporaceae; Candidatus Cytomitobacter
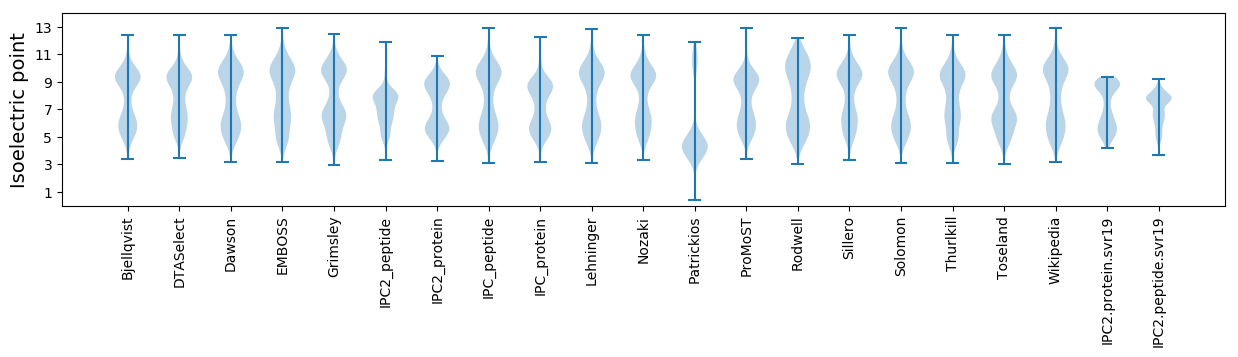
Average proteome isoelectric point is 7.17
Get precalculated fractions of proteins
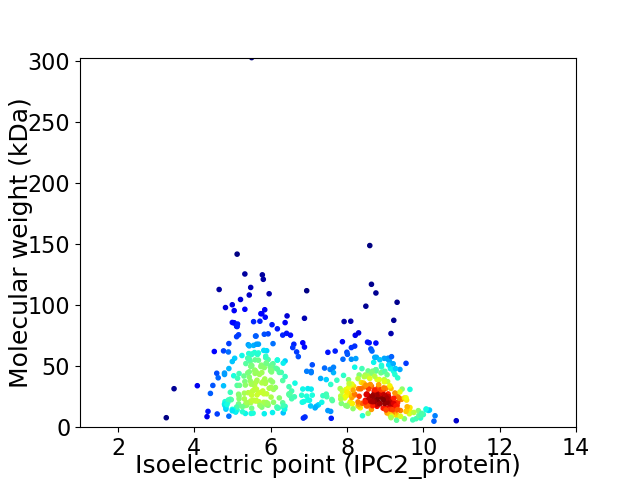
Virtual 2D-PAGE plot for 519 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5C0UIW0|A0A5C0UIW0_9PROT YrdC-like domain-containing protein OS=Candidatus Cytomitobacter primus OX=2066024 GN=FZC34_02785 PE=4 SV=1
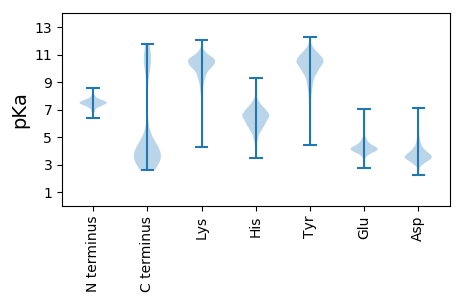
MM1 pKa = 7.28NFISNMIIDD10 pKa = 3.83EE11 pKa = 4.32MFDD14 pKa = 3.61KK15 pKa = 10.78FWNEE19 pKa = 3.21GDD21 pKa = 4.13HH22 pKa = 7.17DD23 pKa = 4.36VSSPVRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84SAEE34 pKa = 3.47SDD36 pKa = 2.88IYY38 pKa = 11.3LRR40 pKa = 11.84FLGAQNSAISIADD53 pKa = 3.34KK54 pKa = 10.95EE55 pKa = 4.65IINKK59 pKa = 8.95LQIDD63 pKa = 4.05DD64 pKa = 5.84RR65 pKa = 11.84ILNDD69 pKa = 3.66DD70 pKa = 4.12NLRR73 pKa = 11.84DD74 pKa = 4.35LIAPFMSQTDD84 pKa = 3.71TVGCLNTHH92 pKa = 6.14ILDD95 pKa = 3.94YY96 pKa = 11.07FDD98 pKa = 6.86DD99 pKa = 4.67IDD101 pKa = 4.03TADD104 pKa = 3.45STVYY108 pKa = 11.31VMLTFMQCFGFKK120 pKa = 10.25TIDD123 pKa = 3.85CFDD126 pKa = 4.14HH127 pKa = 6.58EE128 pKa = 4.9LNIDD132 pKa = 3.3AMIPNRR138 pKa = 11.84EE139 pKa = 3.88FYY141 pKa = 10.78QFNQKK146 pKa = 8.01FTFKK150 pKa = 10.73KK151 pKa = 10.91LEE153 pKa = 4.02ITCNTEE159 pKa = 4.22KK160 pKa = 10.65KK161 pKa = 10.05DD162 pKa = 3.73SKK164 pKa = 11.23PNIEE168 pKa = 4.6DD169 pKa = 4.71DD170 pKa = 3.85INIKK174 pKa = 10.73DD175 pKa = 4.11EE176 pKa = 3.97IVEE179 pKa = 3.97YY180 pKa = 10.75LINTASAQEE189 pKa = 3.81LSFYY193 pKa = 10.84AGKK196 pKa = 10.41DD197 pKa = 3.24CDD199 pKa = 3.57KK200 pKa = 10.27VARR203 pKa = 11.84MVNFVYY209 pKa = 10.48RR210 pKa = 11.84KK211 pKa = 8.71NPEE214 pKa = 3.32MFANLKK220 pKa = 10.26SVGIGVPVYY229 pKa = 11.15GDD231 pKa = 3.66GDD233 pKa = 3.41QDD235 pKa = 3.95LYY237 pKa = 11.35EE238 pKa = 4.14VAVFKK243 pKa = 11.39NDD245 pKa = 3.26VLDD248 pKa = 4.49VLDD251 pKa = 5.23EE252 pKa = 4.66DD253 pKa = 5.34LPCNMHH259 pKa = 5.97MVWMIQGGSDD269 pKa = 4.68SDD271 pKa = 4.66DD272 pKa = 3.21NWVADD277 pKa = 3.79PSDD280 pKa = 4.13YY281 pKa = 10.95IEE283 pKa = 4.74NGFDD287 pKa = 2.89HH288 pKa = 7.39FYY290 pKa = 11.5NEE292 pKa = 5.15FII294 pKa = 4.9
MM1 pKa = 7.28NFISNMIIDD10 pKa = 3.83EE11 pKa = 4.32MFDD14 pKa = 3.61KK15 pKa = 10.78FWNEE19 pKa = 3.21GDD21 pKa = 4.13HH22 pKa = 7.17DD23 pKa = 4.36VSSPVRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84SAEE34 pKa = 3.47SDD36 pKa = 2.88IYY38 pKa = 11.3LRR40 pKa = 11.84FLGAQNSAISIADD53 pKa = 3.34KK54 pKa = 10.95EE55 pKa = 4.65IINKK59 pKa = 8.95LQIDD63 pKa = 4.05DD64 pKa = 5.84RR65 pKa = 11.84ILNDD69 pKa = 3.66DD70 pKa = 4.12NLRR73 pKa = 11.84DD74 pKa = 4.35LIAPFMSQTDD84 pKa = 3.71TVGCLNTHH92 pKa = 6.14ILDD95 pKa = 3.94YY96 pKa = 11.07FDD98 pKa = 6.86DD99 pKa = 4.67IDD101 pKa = 4.03TADD104 pKa = 3.45STVYY108 pKa = 11.31VMLTFMQCFGFKK120 pKa = 10.25TIDD123 pKa = 3.85CFDD126 pKa = 4.14HH127 pKa = 6.58EE128 pKa = 4.9LNIDD132 pKa = 3.3AMIPNRR138 pKa = 11.84EE139 pKa = 3.88FYY141 pKa = 10.78QFNQKK146 pKa = 8.01FTFKK150 pKa = 10.73KK151 pKa = 10.91LEE153 pKa = 4.02ITCNTEE159 pKa = 4.22KK160 pKa = 10.65KK161 pKa = 10.05DD162 pKa = 3.73SKK164 pKa = 11.23PNIEE168 pKa = 4.6DD169 pKa = 4.71DD170 pKa = 3.85INIKK174 pKa = 10.73DD175 pKa = 4.11EE176 pKa = 3.97IVEE179 pKa = 3.97YY180 pKa = 10.75LINTASAQEE189 pKa = 3.81LSFYY193 pKa = 10.84AGKK196 pKa = 10.41DD197 pKa = 3.24CDD199 pKa = 3.57KK200 pKa = 10.27VARR203 pKa = 11.84MVNFVYY209 pKa = 10.48RR210 pKa = 11.84KK211 pKa = 8.71NPEE214 pKa = 3.32MFANLKK220 pKa = 10.26SVGIGVPVYY229 pKa = 11.15GDD231 pKa = 3.66GDD233 pKa = 3.41QDD235 pKa = 3.95LYY237 pKa = 11.35EE238 pKa = 4.14VAVFKK243 pKa = 11.39NDD245 pKa = 3.26VLDD248 pKa = 4.49VLDD251 pKa = 5.23EE252 pKa = 4.66DD253 pKa = 5.34LPCNMHH259 pKa = 5.97MVWMIQGGSDD269 pKa = 4.68SDD271 pKa = 4.66DD272 pKa = 3.21NWVADD277 pKa = 3.79PSDD280 pKa = 4.13YY281 pKa = 10.95IEE283 pKa = 4.74NGFDD287 pKa = 2.89HH288 pKa = 7.39FYY290 pKa = 11.5NEE292 pKa = 5.15FII294 pKa = 4.9
Molecular weight: 33.98 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5C0UF08|A0A5C0UF08_9PROT Uncharacterized protein OS=Candidatus Cytomitobacter primus OX=2066024 GN=FZC34_01810 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.19RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.67GFRR19 pKa = 11.84KK20 pKa = 9.96RR21 pKa = 11.84MSTQDD26 pKa = 2.87GRR28 pKa = 11.84NVMSARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.52GRR39 pKa = 11.84SKK41 pKa = 10.58ISAA44 pKa = 3.55
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.19RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.67GFRR19 pKa = 11.84KK20 pKa = 9.96RR21 pKa = 11.84MSTQDD26 pKa = 2.87GRR28 pKa = 11.84NVMSARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.52GRR39 pKa = 11.84SKK41 pKa = 10.58ISAA44 pKa = 3.55
Molecular weight: 5.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
174856 |
41 |
2712 |
336.9 |
38.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.416 ± 0.091 | 1.35 ± 0.035 |
5.558 ± 0.092 | 6.109 ± 0.127 |
4.508 ± 0.088 | 5.204 ± 0.1 |
2.34 ± 0.044 | 9.669 ± 0.112 |
8.878 ± 0.102 | 9.002 ± 0.158 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.775 ± 0.046 | 7.239 ± 0.118 |
2.78 ± 0.057 | 3.601 ± 0.071 |
3.324 ± 0.076 | 7.871 ± 0.088 |
4.218 ± 0.059 | 5.322 ± 0.088 |
0.876 ± 0.029 | 3.96 ± 0.07 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
