
Nadsonia fulvescens var. elongata DSM 6958
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Saccharomycotina; Saccharomycetes; Saccharomycetales; Saccharomycetales incertae sedis; Nadsonia; Nadsonia fulvescens; Nadsonia fulvescens var. elongata
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
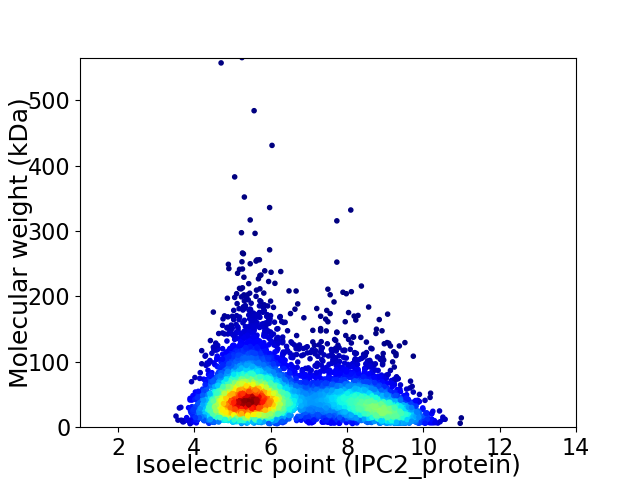
Virtual 2D-PAGE plot for 5636 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1E3PGC5|A0A1E3PGC5_9ASCO MHD domain-containing protein OS=Nadsonia fulvescens var. elongata DSM 6958 OX=857566 GN=NADFUDRAFT_47147 PE=4 SV=1
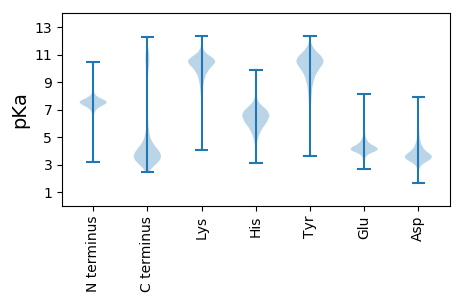
MM1 pKa = 8.05AITITNSRR9 pKa = 11.84PSMADD14 pKa = 3.19YY15 pKa = 10.16TPLAQYY21 pKa = 9.4QASTPASYY29 pKa = 11.21SLDD32 pKa = 3.74EE33 pKa = 4.15NPILHH38 pKa = 6.72FSYY41 pKa = 10.7AGAGLTSSLDD51 pKa = 3.79LPTHH55 pKa = 5.34PTIISKK61 pKa = 9.43TSTATDD67 pKa = 3.17IYY69 pKa = 11.25VLNTDD74 pKa = 3.97LVFWFPNQEE83 pKa = 4.11CGLSVSYY90 pKa = 10.71PDD92 pKa = 4.83IILHH96 pKa = 7.4AIQTQQSPSIYY107 pKa = 10.17LQINFQLGFQGSITEE122 pKa = 4.25SDD124 pKa = 3.36LTDD127 pKa = 5.45DD128 pKa = 3.82YY129 pKa = 11.92DD130 pKa = 4.56NPILEE135 pKa = 4.67LTITMSPEE143 pKa = 3.55DD144 pKa = 4.06QLRR147 pKa = 11.84KK148 pKa = 9.87LYY150 pKa = 10.69SALSHH155 pKa = 6.44CASLHH160 pKa = 6.45PDD162 pKa = 3.61FDD164 pKa = 6.02SDD166 pKa = 4.8DD167 pKa = 3.96QEE169 pKa = 4.18QGAGDD174 pKa = 4.22EE175 pKa = 4.27SDD177 pKa = 4.03SYY179 pKa = 11.79YY180 pKa = 11.38DD181 pKa = 3.57NLEE184 pKa = 4.67DD185 pKa = 4.19EE186 pKa = 5.01DD187 pKa = 5.32AFPEE191 pKa = 3.94DD192 pKa = 4.17HH193 pKa = 6.63QWITAEE199 pKa = 4.96NIDD202 pKa = 3.7SMEE205 pKa = 4.47FGNHH209 pKa = 4.87ASAALEE215 pKa = 4.2VVLDD219 pKa = 4.15EE220 pKa = 5.11PIKK223 pKa = 9.45TGIVRR228 pKa = 11.84RR229 pKa = 11.84RR230 pKa = 11.84EE231 pKa = 4.2SIEE234 pKa = 4.27AEE236 pKa = 3.75ADD238 pKa = 3.35QEE240 pKa = 4.62AGYY243 pKa = 10.88DD244 pKa = 3.64DD245 pKa = 4.73SNNAFNQKK253 pKa = 10.53DD254 pKa = 3.67DD255 pKa = 3.68TVKK258 pKa = 9.34WRR260 pKa = 11.84RR261 pKa = 11.84MEE263 pKa = 3.86
MM1 pKa = 8.05AITITNSRR9 pKa = 11.84PSMADD14 pKa = 3.19YY15 pKa = 10.16TPLAQYY21 pKa = 9.4QASTPASYY29 pKa = 11.21SLDD32 pKa = 3.74EE33 pKa = 4.15NPILHH38 pKa = 6.72FSYY41 pKa = 10.7AGAGLTSSLDD51 pKa = 3.79LPTHH55 pKa = 5.34PTIISKK61 pKa = 9.43TSTATDD67 pKa = 3.17IYY69 pKa = 11.25VLNTDD74 pKa = 3.97LVFWFPNQEE83 pKa = 4.11CGLSVSYY90 pKa = 10.71PDD92 pKa = 4.83IILHH96 pKa = 7.4AIQTQQSPSIYY107 pKa = 10.17LQINFQLGFQGSITEE122 pKa = 4.25SDD124 pKa = 3.36LTDD127 pKa = 5.45DD128 pKa = 3.82YY129 pKa = 11.92DD130 pKa = 4.56NPILEE135 pKa = 4.67LTITMSPEE143 pKa = 3.55DD144 pKa = 4.06QLRR147 pKa = 11.84KK148 pKa = 9.87LYY150 pKa = 10.69SALSHH155 pKa = 6.44CASLHH160 pKa = 6.45PDD162 pKa = 3.61FDD164 pKa = 6.02SDD166 pKa = 4.8DD167 pKa = 3.96QEE169 pKa = 4.18QGAGDD174 pKa = 4.22EE175 pKa = 4.27SDD177 pKa = 4.03SYY179 pKa = 11.79YY180 pKa = 11.38DD181 pKa = 3.57NLEE184 pKa = 4.67DD185 pKa = 4.19EE186 pKa = 5.01DD187 pKa = 5.32AFPEE191 pKa = 3.94DD192 pKa = 4.17HH193 pKa = 6.63QWITAEE199 pKa = 4.96NIDD202 pKa = 3.7SMEE205 pKa = 4.47FGNHH209 pKa = 4.87ASAALEE215 pKa = 4.2VVLDD219 pKa = 4.15EE220 pKa = 5.11PIKK223 pKa = 9.45TGIVRR228 pKa = 11.84RR229 pKa = 11.84RR230 pKa = 11.84EE231 pKa = 4.2SIEE234 pKa = 4.27AEE236 pKa = 3.75ADD238 pKa = 3.35QEE240 pKa = 4.62AGYY243 pKa = 10.88DD244 pKa = 3.64DD245 pKa = 4.73SNNAFNQKK253 pKa = 10.53DD254 pKa = 3.67DD255 pKa = 3.68TVKK258 pKa = 9.34WRR260 pKa = 11.84RR261 pKa = 11.84MEE263 pKa = 3.86
Molecular weight: 29.41 kDa
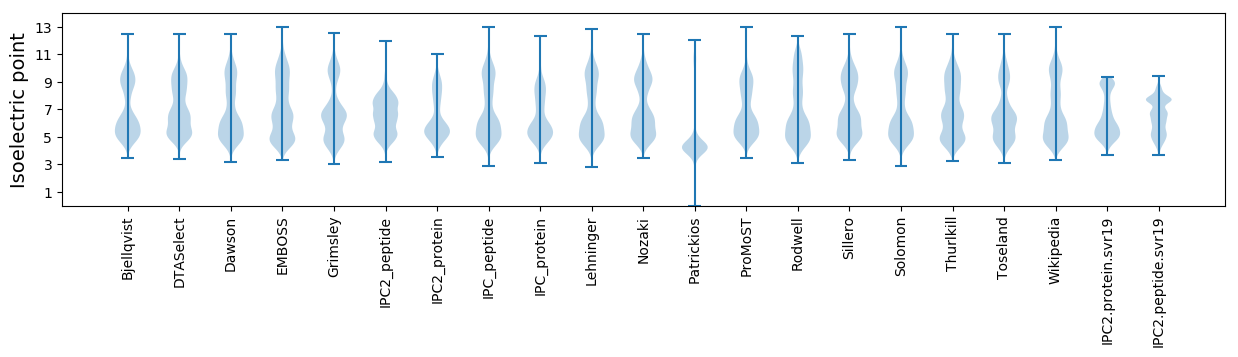
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1E3PKN5|A0A1E3PKN5_9ASCO SET domain-containing protein OS=Nadsonia fulvescens var. elongata DSM 6958 OX=857566 GN=NADFUDRAFT_41974 PE=4 SV=1
MM1 pKa = 7.43FNHH4 pKa = 6.89LSVGLRR10 pKa = 11.84TVPASSRR17 pKa = 11.84FLMGSRR23 pKa = 11.84AFSYY27 pKa = 10.65VPRR30 pKa = 11.84SALTAFRR37 pKa = 11.84AATPSTLNSSLAGRR51 pKa = 11.84EE52 pKa = 4.0STANIAMGSPLLAVPTMGLMSSLAGALQVRR82 pKa = 11.84WKK84 pKa = 10.86SRR86 pKa = 11.84GHH88 pKa = 5.52TFQPNTLKK96 pKa = 10.71RR97 pKa = 11.84KK98 pKa = 9.5RR99 pKa = 11.84KK100 pKa = 9.24FGFLAKK106 pKa = 9.42MRR108 pKa = 11.84AKK110 pKa = 10.18LGRR113 pKa = 11.84VIISRR118 pKa = 11.84RR119 pKa = 11.84RR120 pKa = 11.84AKK122 pKa = 10.25GRR124 pKa = 11.84WYY126 pKa = 10.54LSHH129 pKa = 7.13
MM1 pKa = 7.43FNHH4 pKa = 6.89LSVGLRR10 pKa = 11.84TVPASSRR17 pKa = 11.84FLMGSRR23 pKa = 11.84AFSYY27 pKa = 10.65VPRR30 pKa = 11.84SALTAFRR37 pKa = 11.84AATPSTLNSSLAGRR51 pKa = 11.84EE52 pKa = 4.0STANIAMGSPLLAVPTMGLMSSLAGALQVRR82 pKa = 11.84WKK84 pKa = 10.86SRR86 pKa = 11.84GHH88 pKa = 5.52TFQPNTLKK96 pKa = 10.71RR97 pKa = 11.84KK98 pKa = 9.5RR99 pKa = 11.84KK100 pKa = 9.24FGFLAKK106 pKa = 9.42MRR108 pKa = 11.84AKK110 pKa = 10.18LGRR113 pKa = 11.84VIISRR118 pKa = 11.84RR119 pKa = 11.84RR120 pKa = 11.84AKK122 pKa = 10.25GRR124 pKa = 11.84WYY126 pKa = 10.54LSHH129 pKa = 7.13
Molecular weight: 14.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
2669257 |
49 |
5015 |
473.6 |
52.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.618 ± 0.034 | 1.138 ± 0.011 |
5.804 ± 0.024 | 6.216 ± 0.033 |
4.087 ± 0.021 | 5.6 ± 0.028 |
2.188 ± 0.016 | 6.368 ± 0.021 |
6.19 ± 0.027 | 9.22 ± 0.031 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.023 ± 0.01 | 5.57 ± 0.03 |
4.737 ± 0.027 | 3.924 ± 0.034 |
4.749 ± 0.021 | 9.508 ± 0.05 |
5.938 ± 0.024 | 5.927 ± 0.023 |
1.055 ± 0.011 | 3.134 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
