
Firmicutes bacterium CAG:137
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; environmental samples
Average proteome isoelectric point is 6.28
Get precalculated fractions of proteins
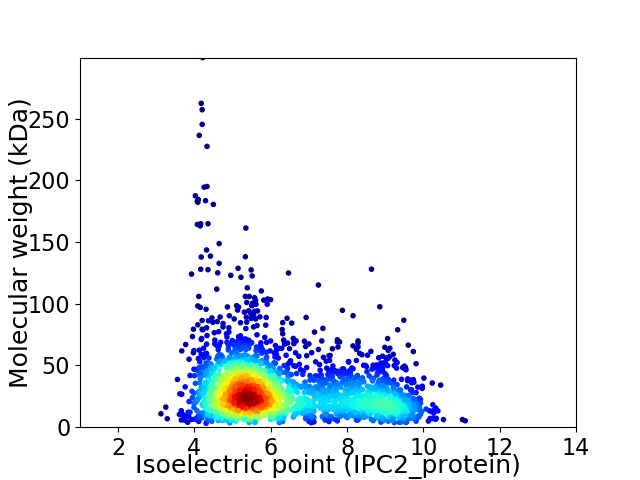
Virtual 2D-PAGE plot for 2347 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R6GXH3|R6GXH3_9FIRM Undecaprenyl-diphosphatase OS=Firmicutes bacterium CAG:137 OX=1263004 GN=uppP PE=3 SV=1
MM1 pKa = 7.27LTNHH5 pKa = 6.22HH6 pKa = 6.88VIDD9 pKa = 4.81GAQSITVTLSDD20 pKa = 3.43GTEE23 pKa = 3.89YY24 pKa = 10.06TAQLVGSDD32 pKa = 3.73SVSDD36 pKa = 3.78VALLKK41 pKa = 10.08IDD43 pKa = 4.26GQDD46 pKa = 3.4LPAVSIGNSDD56 pKa = 3.92DD57 pKa = 5.52LEE59 pKa = 5.13VGDD62 pKa = 4.27QVAAIGNPLGEE73 pKa = 4.4LTSTQTVGYY82 pKa = 9.78VSAKK86 pKa = 9.94DD87 pKa = 3.36RR88 pKa = 11.84SVNTDD93 pKa = 2.78GTIINMLQTDD103 pKa = 3.9AAINSGNSGGPLFNMNGEE121 pKa = 4.47VVGITTAKK129 pKa = 10.21YY130 pKa = 10.3SGSTSSGASIEE141 pKa = 4.64GIGFAIPINDD151 pKa = 3.39VMKK154 pKa = 10.89LVDD157 pKa = 4.89DD158 pKa = 4.11LMEE161 pKa = 4.17YY162 pKa = 10.99GYY164 pKa = 8.53ITGQAYY170 pKa = 10.59LGVTINSKK178 pKa = 10.86DD179 pKa = 3.57LDD181 pKa = 3.95ASTAAAYY188 pKa = 8.14GLPVGARR195 pKa = 11.84VEE197 pKa = 4.54SVTEE201 pKa = 4.2GSCAQKK207 pKa = 10.89AGLQAGDD214 pKa = 4.37IITALGDD221 pKa = 3.67QKK223 pKa = 11.51VSGYY227 pKa = 10.62SDD229 pKa = 3.64LVYY232 pKa = 11.08ALRR235 pKa = 11.84NFSAGDD241 pKa = 3.34ATTVSVYY248 pKa = 10.44RR249 pKa = 11.84AGQEE253 pKa = 3.65LTLQITFDD261 pKa = 3.83EE262 pKa = 4.63KK263 pKa = 10.58TADD266 pKa = 3.6TVTGPVVTTEE276 pKa = 3.99PTEE279 pKa = 3.91ATGDD283 pKa = 3.97GEE285 pKa = 4.83QPSQDD290 pKa = 4.34EE291 pKa = 4.4IPEE294 pKa = 4.34SGSYY298 pKa = 9.66EE299 pKa = 3.61DD300 pKa = 3.75WYY302 pKa = 10.45DD303 pKa = 3.23YY304 pKa = 10.76FYY306 pKa = 11.31RR307 pKa = 11.84FFGGNGNN314 pKa = 3.64
MM1 pKa = 7.27LTNHH5 pKa = 6.22HH6 pKa = 6.88VIDD9 pKa = 4.81GAQSITVTLSDD20 pKa = 3.43GTEE23 pKa = 3.89YY24 pKa = 10.06TAQLVGSDD32 pKa = 3.73SVSDD36 pKa = 3.78VALLKK41 pKa = 10.08IDD43 pKa = 4.26GQDD46 pKa = 3.4LPAVSIGNSDD56 pKa = 3.92DD57 pKa = 5.52LEE59 pKa = 5.13VGDD62 pKa = 4.27QVAAIGNPLGEE73 pKa = 4.4LTSTQTVGYY82 pKa = 9.78VSAKK86 pKa = 9.94DD87 pKa = 3.36RR88 pKa = 11.84SVNTDD93 pKa = 2.78GTIINMLQTDD103 pKa = 3.9AAINSGNSGGPLFNMNGEE121 pKa = 4.47VVGITTAKK129 pKa = 10.21YY130 pKa = 10.3SGSTSSGASIEE141 pKa = 4.64GIGFAIPINDD151 pKa = 3.39VMKK154 pKa = 10.89LVDD157 pKa = 4.89DD158 pKa = 4.11LMEE161 pKa = 4.17YY162 pKa = 10.99GYY164 pKa = 8.53ITGQAYY170 pKa = 10.59LGVTINSKK178 pKa = 10.86DD179 pKa = 3.57LDD181 pKa = 3.95ASTAAAYY188 pKa = 8.14GLPVGARR195 pKa = 11.84VEE197 pKa = 4.54SVTEE201 pKa = 4.2GSCAQKK207 pKa = 10.89AGLQAGDD214 pKa = 4.37IITALGDD221 pKa = 3.67QKK223 pKa = 11.51VSGYY227 pKa = 10.62SDD229 pKa = 3.64LVYY232 pKa = 11.08ALRR235 pKa = 11.84NFSAGDD241 pKa = 3.34ATTVSVYY248 pKa = 10.44RR249 pKa = 11.84AGQEE253 pKa = 3.65LTLQITFDD261 pKa = 3.83EE262 pKa = 4.63KK263 pKa = 10.58TADD266 pKa = 3.6TVTGPVVTTEE276 pKa = 3.99PTEE279 pKa = 3.91ATGDD283 pKa = 3.97GEE285 pKa = 4.83QPSQDD290 pKa = 4.34EE291 pKa = 4.4IPEE294 pKa = 4.34SGSYY298 pKa = 9.66EE299 pKa = 3.61DD300 pKa = 3.75WYY302 pKa = 10.45DD303 pKa = 3.23YY304 pKa = 10.76FYY306 pKa = 11.31RR307 pKa = 11.84FFGGNGNN314 pKa = 3.64
Molecular weight: 32.64 kDa
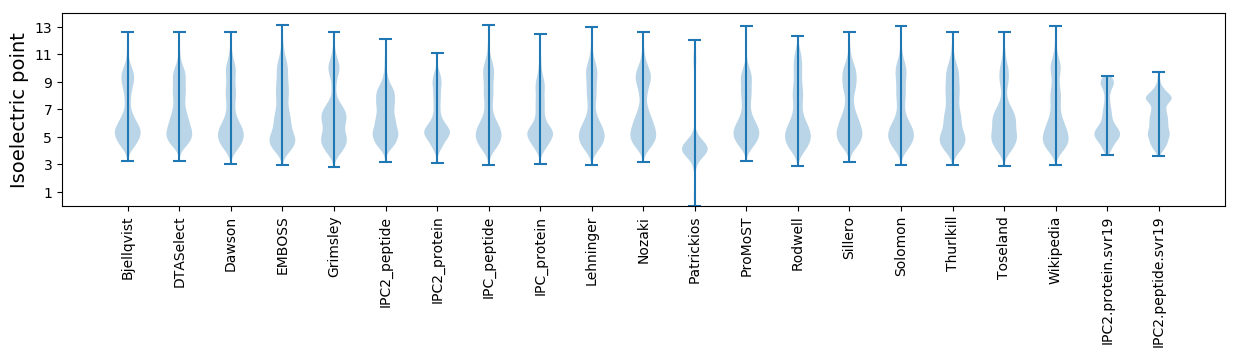
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R6H7T2|R6H7T2_9FIRM CRISPR system Cms protein Csm5 OS=Firmicutes bacterium CAG:137 OX=1263004 GN=BN490_00812 PE=3 SV=1
MM1 pKa = 7.45LRR3 pKa = 11.84TYY5 pKa = 9.82QPKK8 pKa = 9.37KK9 pKa = 7.72RR10 pKa = 11.84HH11 pKa = 5.14RR12 pKa = 11.84AKK14 pKa = 10.52VHH16 pKa = 5.63GFRR19 pKa = 11.84QRR21 pKa = 11.84MSTTNGRR28 pKa = 11.84KK29 pKa = 8.65VLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.99GRR39 pKa = 11.84KK40 pKa = 8.67VLSAA44 pKa = 4.05
MM1 pKa = 7.45LRR3 pKa = 11.84TYY5 pKa = 9.82QPKK8 pKa = 9.37KK9 pKa = 7.72RR10 pKa = 11.84HH11 pKa = 5.14RR12 pKa = 11.84AKK14 pKa = 10.52VHH16 pKa = 5.63GFRR19 pKa = 11.84QRR21 pKa = 11.84MSTTNGRR28 pKa = 11.84KK29 pKa = 8.65VLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.99GRR39 pKa = 11.84KK40 pKa = 8.67VLSAA44 pKa = 4.05
Molecular weight: 5.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
691874 |
29 |
2786 |
294.8 |
32.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.386 ± 0.058 | 1.784 ± 0.024 |
5.175 ± 0.045 | 6.442 ± 0.057 |
3.71 ± 0.034 | 8.375 ± 0.068 |
2.139 ± 0.037 | 5.208 ± 0.045 |
4.09 ± 0.05 | 10.365 ± 0.071 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.625 ± 0.027 | 3.236 ± 0.038 |
4.432 ± 0.038 | 4.205 ± 0.037 |
5.73 ± 0.064 | 5.555 ± 0.042 |
5.592 ± 0.063 | 7.297 ± 0.052 |
1.097 ± 0.021 | 3.541 ± 0.046 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
