
Cafeteria roenbergensis (Marine flagellate)
Taxonomy: cellular organisms; Eukaryota; Sar; Stramenopiles; Bigyra; Opalozoa; Bicosoecida; Cafeteriaceae; Cafeteria
Average proteome isoelectric point is 6.25
Get precalculated fractions of proteins
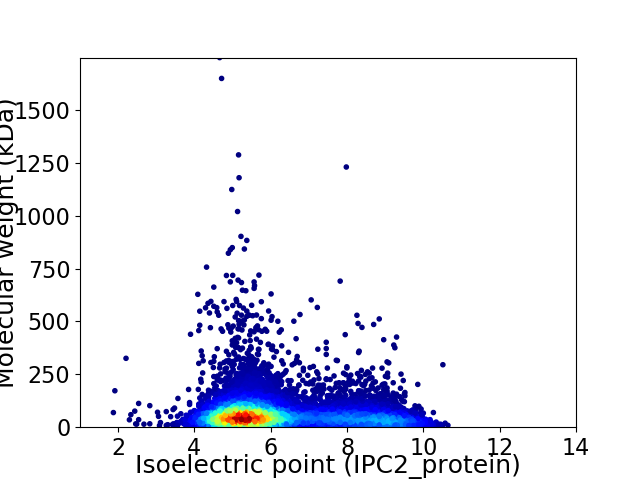
Virtual 2D-PAGE plot for 8530 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
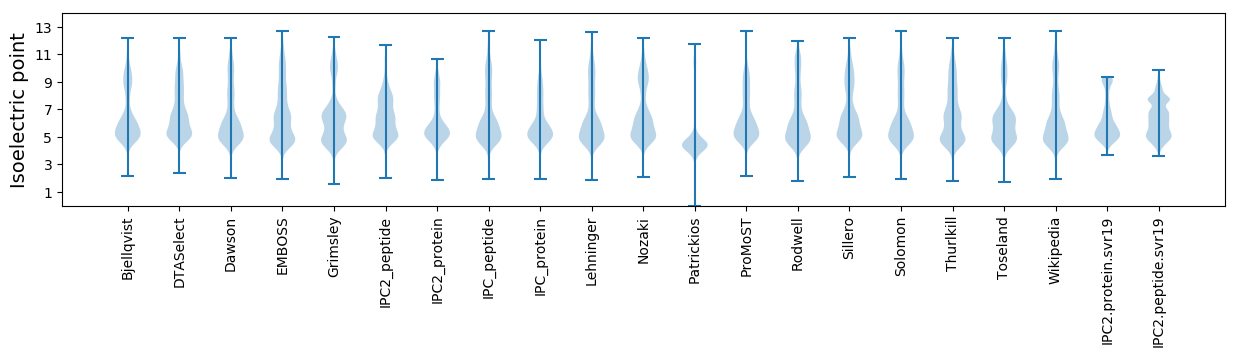
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5A8CVE5|A0A5A8CVE5_CAFRO Uncharacterized protein OS=Cafeteria roenbergensis OX=33653 GN=FNF29_01199 PE=4 SV=1
MM1 pKa = 7.38NAFDD5 pKa = 6.33GPIHH9 pKa = 5.94WQQIATSGAYY19 pKa = 10.1VFVTILGLVTIPHH32 pKa = 6.23MFSNSEE38 pKa = 4.0LPLALSLTIYY48 pKa = 10.79LLLAAVCVASWLYY61 pKa = 10.49IEE63 pKa = 5.09LTDD66 pKa = 3.79PSMPGGVACPCMKK79 pKa = 9.72RR80 pKa = 11.84TQSSQSNCSLCRR92 pKa = 11.84KK93 pKa = 8.95PVVGLDD99 pKa = 2.98HH100 pKa = 7.13HH101 pKa = 7.24CYY103 pKa = 10.19FLNTCIGKK111 pKa = 9.92RR112 pKa = 11.84NYY114 pKa = 10.88AHH116 pKa = 7.0FFALSLCSTLQMAAQVVFSVLGMTVWIEE144 pKa = 3.88RR145 pKa = 11.84GNDD148 pKa = 2.9NVLYY152 pKa = 9.65YY153 pKa = 10.96VLVGLQALIAAVAGGATCSLLGFHH177 pKa = 7.22VYY179 pKa = 10.53LLWIGMGTYY188 pKa = 10.68DD189 pKa = 5.07WMWAKK194 pKa = 10.03QDD196 pKa = 3.33KK197 pKa = 10.26AAEE200 pKa = 4.15KK201 pKa = 10.53VRR203 pKa = 11.84RR204 pKa = 11.84EE205 pKa = 3.69NQARR209 pKa = 11.84FEE211 pKa = 4.43AEE213 pKa = 3.67QQARR217 pKa = 11.84LQLEE221 pKa = 4.31RR222 pKa = 11.84EE223 pKa = 4.17RR224 pKa = 11.84QQAHH228 pKa = 5.9GAAPAMPAGRR238 pKa = 11.84VAVTVEE244 pKa = 3.97SHH246 pKa = 6.38PVAAAAGAAAAAGVGGDD263 pKa = 3.81SGGGLYY269 pKa = 10.52AGFEE273 pKa = 4.28PEE275 pKa = 3.76TDD277 pKa = 3.48EE278 pKa = 5.48EE279 pKa = 4.52EE280 pKa = 4.7GGTAGGADD288 pKa = 3.77TASSPGDD295 pKa = 3.43DD296 pKa = 3.91YY297 pKa = 12.1GGVDD301 pKa = 3.81AGEE304 pKa = 4.64GGAGAEE310 pKa = 4.21SQDD313 pKa = 4.05VIGMEE318 pKa = 4.28YY319 pKa = 10.52ADD321 pKa = 4.19EE322 pKa = 4.8AGGVYY327 pKa = 10.56DD328 pKa = 5.65DD329 pKa = 5.35DD330 pKa = 4.83AAGAYY335 pKa = 10.27DD336 pKa = 4.76DD337 pKa = 5.41DD338 pKa = 4.69AAGGAFDD345 pKa = 5.09DD346 pKa = 4.81EE347 pKa = 4.52GRR349 pKa = 11.84EE350 pKa = 3.94GDD352 pKa = 3.78GFGDD356 pKa = 3.85EE357 pKa = 4.53EE358 pKa = 5.67DD359 pKa = 4.1EE360 pKa = 4.5ADD362 pKa = 3.46HH363 pKa = 6.82EE364 pKa = 4.5RR365 pKa = 11.84VQRR368 pKa = 11.84VPGASSAGSTGAPSGDD384 pKa = 3.82SAPGSPTQAPTEE396 pKa = 4.09EE397 pKa = 4.69GGAEE401 pKa = 3.74QGAEE405 pKa = 4.06AEE407 pKa = 4.55EE408 pKa = 4.52AGSPSDD414 pKa = 4.28AGGEE418 pKa = 3.93AHH420 pKa = 6.1GQEE423 pKa = 4.3GAEE426 pKa = 3.97QEE428 pKa = 4.37EE429 pKa = 4.81AEE431 pKa = 4.43DD432 pKa = 3.89AASGEE437 pKa = 4.38AASGSQAAGEE447 pKa = 4.06PAEE450 pKa = 4.34TEE452 pKa = 3.99ATAEE456 pKa = 4.08ATGPEE461 pKa = 3.97AAAIAQEE468 pKa = 4.27EE469 pKa = 4.39SDD471 pKa = 5.02ADD473 pKa = 3.7AADD476 pKa = 4.22AGDD479 pKa = 4.8DD480 pKa = 3.75AVADD484 pKa = 4.39AGDD487 pKa = 4.04DD488 pKa = 3.71AGDD491 pKa = 3.92AAVDD495 pKa = 4.26DD496 pKa = 5.16AGADD500 pKa = 3.48AGDD503 pKa = 4.04DD504 pKa = 3.23AGAPGEE510 pKa = 4.31EE511 pKa = 4.43EE512 pKa = 4.18GLADD516 pKa = 4.6GAADD520 pKa = 3.72AAAPAAPVAAPEE532 pKa = 4.62GGPEE536 pKa = 4.25GDD538 pKa = 4.05SCDD541 pKa = 3.48STGRR545 pKa = 11.84PGSAGEE551 pKa = 4.27LQPEE555 pKa = 4.49TATPRR560 pKa = 11.84PGEE563 pKa = 3.9AA564 pKa = 3.76
MM1 pKa = 7.38NAFDD5 pKa = 6.33GPIHH9 pKa = 5.94WQQIATSGAYY19 pKa = 10.1VFVTILGLVTIPHH32 pKa = 6.23MFSNSEE38 pKa = 4.0LPLALSLTIYY48 pKa = 10.79LLLAAVCVASWLYY61 pKa = 10.49IEE63 pKa = 5.09LTDD66 pKa = 3.79PSMPGGVACPCMKK79 pKa = 9.72RR80 pKa = 11.84TQSSQSNCSLCRR92 pKa = 11.84KK93 pKa = 8.95PVVGLDD99 pKa = 2.98HH100 pKa = 7.13HH101 pKa = 7.24CYY103 pKa = 10.19FLNTCIGKK111 pKa = 9.92RR112 pKa = 11.84NYY114 pKa = 10.88AHH116 pKa = 7.0FFALSLCSTLQMAAQVVFSVLGMTVWIEE144 pKa = 3.88RR145 pKa = 11.84GNDD148 pKa = 2.9NVLYY152 pKa = 9.65YY153 pKa = 10.96VLVGLQALIAAVAGGATCSLLGFHH177 pKa = 7.22VYY179 pKa = 10.53LLWIGMGTYY188 pKa = 10.68DD189 pKa = 5.07WMWAKK194 pKa = 10.03QDD196 pKa = 3.33KK197 pKa = 10.26AAEE200 pKa = 4.15KK201 pKa = 10.53VRR203 pKa = 11.84RR204 pKa = 11.84EE205 pKa = 3.69NQARR209 pKa = 11.84FEE211 pKa = 4.43AEE213 pKa = 3.67QQARR217 pKa = 11.84LQLEE221 pKa = 4.31RR222 pKa = 11.84EE223 pKa = 4.17RR224 pKa = 11.84QQAHH228 pKa = 5.9GAAPAMPAGRR238 pKa = 11.84VAVTVEE244 pKa = 3.97SHH246 pKa = 6.38PVAAAAGAAAAAGVGGDD263 pKa = 3.81SGGGLYY269 pKa = 10.52AGFEE273 pKa = 4.28PEE275 pKa = 3.76TDD277 pKa = 3.48EE278 pKa = 5.48EE279 pKa = 4.52EE280 pKa = 4.7GGTAGGADD288 pKa = 3.77TASSPGDD295 pKa = 3.43DD296 pKa = 3.91YY297 pKa = 12.1GGVDD301 pKa = 3.81AGEE304 pKa = 4.64GGAGAEE310 pKa = 4.21SQDD313 pKa = 4.05VIGMEE318 pKa = 4.28YY319 pKa = 10.52ADD321 pKa = 4.19EE322 pKa = 4.8AGGVYY327 pKa = 10.56DD328 pKa = 5.65DD329 pKa = 5.35DD330 pKa = 4.83AAGAYY335 pKa = 10.27DD336 pKa = 4.76DD337 pKa = 5.41DD338 pKa = 4.69AAGGAFDD345 pKa = 5.09DD346 pKa = 4.81EE347 pKa = 4.52GRR349 pKa = 11.84EE350 pKa = 3.94GDD352 pKa = 3.78GFGDD356 pKa = 3.85EE357 pKa = 4.53EE358 pKa = 5.67DD359 pKa = 4.1EE360 pKa = 4.5ADD362 pKa = 3.46HH363 pKa = 6.82EE364 pKa = 4.5RR365 pKa = 11.84VQRR368 pKa = 11.84VPGASSAGSTGAPSGDD384 pKa = 3.82SAPGSPTQAPTEE396 pKa = 4.09EE397 pKa = 4.69GGAEE401 pKa = 3.74QGAEE405 pKa = 4.06AEE407 pKa = 4.55EE408 pKa = 4.52AGSPSDD414 pKa = 4.28AGGEE418 pKa = 3.93AHH420 pKa = 6.1GQEE423 pKa = 4.3GAEE426 pKa = 3.97QEE428 pKa = 4.37EE429 pKa = 4.81AEE431 pKa = 4.43DD432 pKa = 3.89AASGEE437 pKa = 4.38AASGSQAAGEE447 pKa = 4.06PAEE450 pKa = 4.34TEE452 pKa = 3.99ATAEE456 pKa = 4.08ATGPEE461 pKa = 3.97AAAIAQEE468 pKa = 4.27EE469 pKa = 4.39SDD471 pKa = 5.02ADD473 pKa = 3.7AADD476 pKa = 4.22AGDD479 pKa = 4.8DD480 pKa = 3.75AVADD484 pKa = 4.39AGDD487 pKa = 4.04DD488 pKa = 3.71AGDD491 pKa = 3.92AAVDD495 pKa = 4.26DD496 pKa = 5.16AGADD500 pKa = 3.48AGDD503 pKa = 4.04DD504 pKa = 3.23AGAPGEE510 pKa = 4.31EE511 pKa = 4.43EE512 pKa = 4.18GLADD516 pKa = 4.6GAADD520 pKa = 3.72AAAPAAPVAAPEE532 pKa = 4.62GGPEE536 pKa = 4.25GDD538 pKa = 4.05SCDD541 pKa = 3.48STGRR545 pKa = 11.84PGSAGEE551 pKa = 4.27LQPEE555 pKa = 4.49TATPRR560 pKa = 11.84PGEE563 pKa = 3.9AA564 pKa = 3.76
Molecular weight: 56.68 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5A8CW52|A0A5A8CW52_CAFRO Uncharacterized protein OS=Cafeteria roenbergensis OX=33653 GN=FNF29_00708 PE=4 SV=1
MM1 pKa = 7.51AALGVVVFFIVLVLPMLAITFDD23 pKa = 3.36AARR26 pKa = 11.84RR27 pKa = 11.84NSPRR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84LDD35 pKa = 3.33PGTTLVVLMVLSMTPHH51 pKa = 6.33AMGLALRR58 pKa = 11.84EE59 pKa = 4.16ALLATARR66 pKa = 11.84GSLIRR71 pKa = 11.84LVVTQATVLFCTALTWVMEE90 pKa = 4.42LAAQAMTPIEE100 pKa = 4.56AEE102 pKa = 4.04RR103 pKa = 11.84QPLLVAADD111 pKa = 3.52VMLTVFLNLVTAQEE125 pKa = 4.24LAVGTVEE132 pKa = 4.81FWLTLAVRR140 pKa = 11.84CAGLLFMDD148 pKa = 3.62TRR150 pKa = 11.84LWEE153 pKa = 4.41DD154 pKa = 3.5VQSLFTNKK162 pKa = 9.29RR163 pKa = 11.84RR164 pKa = 11.84WFRR167 pKa = 11.84FVTARR172 pKa = 11.84LGWLAVQRR180 pKa = 11.84ADD182 pKa = 3.22RR183 pKa = 11.84VLVSEE188 pKa = 4.06QLALVISAAYY198 pKa = 8.28LTGGAAGALAGFVSAQEE215 pKa = 3.8ATAAEE220 pKa = 4.32NPRR223 pKa = 11.84EE224 pKa = 4.07VVGRR228 pKa = 11.84VTAFAVIFVGISIMVPVVRR247 pKa = 11.84YY248 pKa = 7.69LTYY251 pKa = 11.15AKK253 pKa = 10.33LARR256 pKa = 11.84ARR258 pKa = 11.84LKK260 pKa = 10.62SAAVSILGSCTVLNLVKK277 pKa = 10.36DD278 pKa = 3.53RR279 pKa = 11.84HH280 pKa = 5.27AAHH283 pKa = 7.53RR284 pKa = 11.84IAGVAAAGGSFFGDD298 pKa = 3.35DD299 pKa = 3.53CKK301 pKa = 10.85RR302 pKa = 11.84WGVASRR308 pKa = 11.84EE309 pKa = 4.33SLSDD313 pKa = 3.4RR314 pKa = 11.84SPPFASVRR322 pKa = 11.84QHH324 pKa = 6.25RR325 pKa = 11.84GSFVPYY331 pKa = 10.27VFKK334 pKa = 10.75EE335 pKa = 3.88WVNGVRR341 pKa = 11.84QSRR344 pKa = 11.84GKK346 pKa = 9.07GCARR350 pKa = 11.84PRR352 pKa = 11.84CIRR355 pKa = 11.84VGILCFGDD363 pKa = 3.44ARR365 pKa = 11.84SSGQHH370 pKa = 3.83WVAFQTEE377 pKa = 4.25YY378 pKa = 11.54AFAVMLFAALWGSWASSVALGVKK401 pKa = 10.09AFSS404 pKa = 3.5
MM1 pKa = 7.51AALGVVVFFIVLVLPMLAITFDD23 pKa = 3.36AARR26 pKa = 11.84RR27 pKa = 11.84NSPRR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84LDD35 pKa = 3.33PGTTLVVLMVLSMTPHH51 pKa = 6.33AMGLALRR58 pKa = 11.84EE59 pKa = 4.16ALLATARR66 pKa = 11.84GSLIRR71 pKa = 11.84LVVTQATVLFCTALTWVMEE90 pKa = 4.42LAAQAMTPIEE100 pKa = 4.56AEE102 pKa = 4.04RR103 pKa = 11.84QPLLVAADD111 pKa = 3.52VMLTVFLNLVTAQEE125 pKa = 4.24LAVGTVEE132 pKa = 4.81FWLTLAVRR140 pKa = 11.84CAGLLFMDD148 pKa = 3.62TRR150 pKa = 11.84LWEE153 pKa = 4.41DD154 pKa = 3.5VQSLFTNKK162 pKa = 9.29RR163 pKa = 11.84RR164 pKa = 11.84WFRR167 pKa = 11.84FVTARR172 pKa = 11.84LGWLAVQRR180 pKa = 11.84ADD182 pKa = 3.22RR183 pKa = 11.84VLVSEE188 pKa = 4.06QLALVISAAYY198 pKa = 8.28LTGGAAGALAGFVSAQEE215 pKa = 3.8ATAAEE220 pKa = 4.32NPRR223 pKa = 11.84EE224 pKa = 4.07VVGRR228 pKa = 11.84VTAFAVIFVGISIMVPVVRR247 pKa = 11.84YY248 pKa = 7.69LTYY251 pKa = 11.15AKK253 pKa = 10.33LARR256 pKa = 11.84ARR258 pKa = 11.84LKK260 pKa = 10.62SAAVSILGSCTVLNLVKK277 pKa = 10.36DD278 pKa = 3.53RR279 pKa = 11.84HH280 pKa = 5.27AAHH283 pKa = 7.53RR284 pKa = 11.84IAGVAAAGGSFFGDD298 pKa = 3.35DD299 pKa = 3.53CKK301 pKa = 10.85RR302 pKa = 11.84WGVASRR308 pKa = 11.84EE309 pKa = 4.33SLSDD313 pKa = 3.4RR314 pKa = 11.84SPPFASVRR322 pKa = 11.84QHH324 pKa = 6.25RR325 pKa = 11.84GSFVPYY331 pKa = 10.27VFKK334 pKa = 10.75EE335 pKa = 3.88WVNGVRR341 pKa = 11.84QSRR344 pKa = 11.84GKK346 pKa = 9.07GCARR350 pKa = 11.84PRR352 pKa = 11.84CIRR355 pKa = 11.84VGILCFGDD363 pKa = 3.44ARR365 pKa = 11.84SSGQHH370 pKa = 3.83WVAFQTEE377 pKa = 4.25YY378 pKa = 11.54AFAVMLFAALWGSWASSVALGVKK401 pKa = 10.09AFSS404 pKa = 3.5
Molecular weight: 43.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
5899624 |
66 |
17662 |
691.6 |
72.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
17.239 ± 0.069 | 1.257 ± 0.01 |
5.696 ± 0.022 | 5.814 ± 0.031 |
2.71 ± 0.017 | 9.457 ± 0.036 |
1.863 ± 0.012 | 2.439 ± 0.019 |
2.589 ± 0.023 | 8.728 ± 0.028 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.896 ± 0.012 | 1.613 ± 0.015 |
5.825 ± 0.026 | 2.824 ± 0.012 |
7.383 ± 0.03 | 8.051 ± 0.029 |
4.787 ± 0.027 | 7.265 ± 0.035 |
1.271 ± 0.009 | 1.293 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
