
Porcine rotavirus A
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Rotavirus; Rotavirus A
Average proteome isoelectric point is 6.01
Get precalculated fractions of proteins
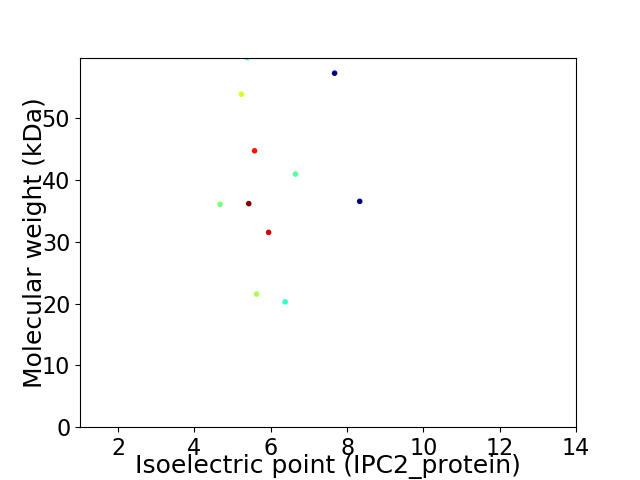
Virtual 2D-PAGE plot for 11 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
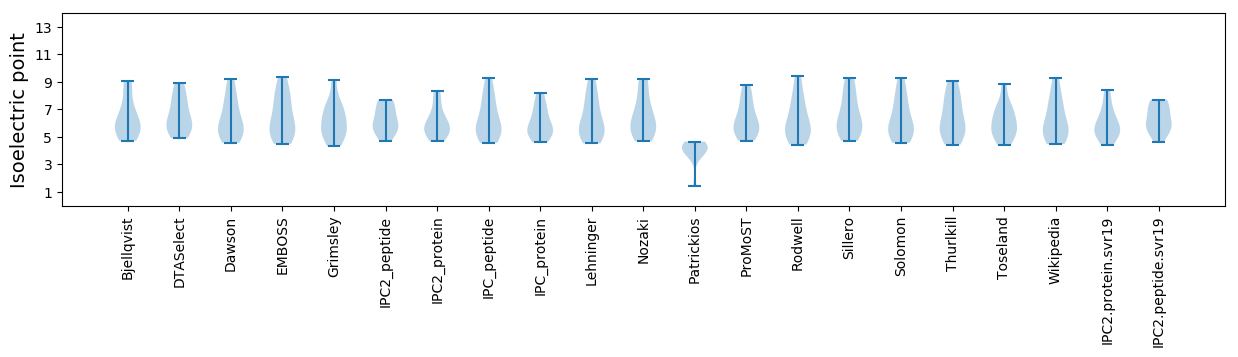
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q06B31|Q06B31_9REOV Non-structural protein 5 OS=Porcine rotavirus A OX=10967 GN=NSP5 PE=3 SV=1
MM1 pKa = 7.26YY2 pKa = 10.2GIEE5 pKa = 4.22YY6 pKa = 7.59TTILTFLISLIFINYY21 pKa = 8.0ILKK24 pKa = 10.48SVTRR28 pKa = 11.84TMDD31 pKa = 3.3FVIYY35 pKa = 9.85RR36 pKa = 11.84SLFIIVMLAPFIKK49 pKa = 8.7TQNYY53 pKa = 9.32GINLPITGSMDD64 pKa = 3.49TPYY67 pKa = 10.26MNSTASEE74 pKa = 4.34TFLTSTLCLYY84 pKa = 10.23YY85 pKa = 10.01PNEE88 pKa = 4.23AARR91 pKa = 11.84QIADD95 pKa = 4.6DD96 pKa = 3.6KK97 pKa = 11.4WKK99 pKa = 9.31DD100 pKa = 3.44TLSQLFLTKK109 pKa = 10.05GWPTGSVYY117 pKa = 10.44FKK119 pKa = 10.13EE120 pKa = 4.36YY121 pKa = 10.91VDD123 pKa = 3.34IASFSVDD130 pKa = 3.05PQLYY134 pKa = 8.88CDD136 pKa = 3.82YY137 pKa = 10.47NIVIMKK143 pKa = 10.15YY144 pKa = 10.48DD145 pKa = 3.49VDD147 pKa = 4.15SQLDD151 pKa = 3.66MSEE154 pKa = 4.1LADD157 pKa = 5.13LILNEE162 pKa = 4.24WLCNPMDD169 pKa = 3.6IALYY173 pKa = 10.23YY174 pKa = 9.03YY175 pKa = 10.41QQTDD179 pKa = 3.57EE180 pKa = 4.17ANKK183 pKa = 8.13WISMGTSCTIKK194 pKa = 10.29VCPLNTQTLGVGCLTTDD211 pKa = 3.05PTTFEE216 pKa = 3.89EE217 pKa = 4.17VAYY220 pKa = 10.34AEE222 pKa = 4.51KK223 pKa = 10.68LAITDD228 pKa = 3.46VVDD231 pKa = 4.79GVDD234 pKa = 3.32HH235 pKa = 6.79KK236 pKa = 11.72LDD238 pKa = 3.66VTTTTCTIRR247 pKa = 11.84NCKK250 pKa = 9.83KK251 pKa = 10.18LGPRR255 pKa = 11.84EE256 pKa = 3.99NVAVIQIGGSNILDD270 pKa = 3.33ITADD274 pKa = 3.68PTTAPQTEE282 pKa = 3.66RR283 pKa = 11.84MMRR286 pKa = 11.84INWKK290 pKa = 9.54KK291 pKa = 7.26WWQVFYY297 pKa = 10.66TIVDD301 pKa = 3.57YY302 pKa = 11.59VNQIVQAMSKK312 pKa = 10.16RR313 pKa = 11.84SRR315 pKa = 11.84SS316 pKa = 3.46
MM1 pKa = 7.26YY2 pKa = 10.2GIEE5 pKa = 4.22YY6 pKa = 7.59TTILTFLISLIFINYY21 pKa = 8.0ILKK24 pKa = 10.48SVTRR28 pKa = 11.84TMDD31 pKa = 3.3FVIYY35 pKa = 9.85RR36 pKa = 11.84SLFIIVMLAPFIKK49 pKa = 8.7TQNYY53 pKa = 9.32GINLPITGSMDD64 pKa = 3.49TPYY67 pKa = 10.26MNSTASEE74 pKa = 4.34TFLTSTLCLYY84 pKa = 10.23YY85 pKa = 10.01PNEE88 pKa = 4.23AARR91 pKa = 11.84QIADD95 pKa = 4.6DD96 pKa = 3.6KK97 pKa = 11.4WKK99 pKa = 9.31DD100 pKa = 3.44TLSQLFLTKK109 pKa = 10.05GWPTGSVYY117 pKa = 10.44FKK119 pKa = 10.13EE120 pKa = 4.36YY121 pKa = 10.91VDD123 pKa = 3.34IASFSVDD130 pKa = 3.05PQLYY134 pKa = 8.88CDD136 pKa = 3.82YY137 pKa = 10.47NIVIMKK143 pKa = 10.15YY144 pKa = 10.48DD145 pKa = 3.49VDD147 pKa = 4.15SQLDD151 pKa = 3.66MSEE154 pKa = 4.1LADD157 pKa = 5.13LILNEE162 pKa = 4.24WLCNPMDD169 pKa = 3.6IALYY173 pKa = 10.23YY174 pKa = 9.03YY175 pKa = 10.41QQTDD179 pKa = 3.57EE180 pKa = 4.17ANKK183 pKa = 8.13WISMGTSCTIKK194 pKa = 10.29VCPLNTQTLGVGCLTTDD211 pKa = 3.05PTTFEE216 pKa = 3.89EE217 pKa = 4.17VAYY220 pKa = 10.34AEE222 pKa = 4.51KK223 pKa = 10.68LAITDD228 pKa = 3.46VVDD231 pKa = 4.79GVDD234 pKa = 3.32HH235 pKa = 6.79KK236 pKa = 11.72LDD238 pKa = 3.66VTTTTCTIRR247 pKa = 11.84NCKK250 pKa = 9.83KK251 pKa = 10.18LGPRR255 pKa = 11.84EE256 pKa = 3.99NVAVIQIGGSNILDD270 pKa = 3.33ITADD274 pKa = 3.68PTTAPQTEE282 pKa = 3.66RR283 pKa = 11.84MMRR286 pKa = 11.84INWKK290 pKa = 9.54KK291 pKa = 7.26WWQVFYY297 pKa = 10.66TIVDD301 pKa = 3.57YY302 pKa = 11.59VNQIVQAMSKK312 pKa = 10.16RR313 pKa = 11.84SRR315 pKa = 11.84SS316 pKa = 3.46
Molecular weight: 36.09 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D3Q266|A0A0D3Q266_9REOV Non-structural protein 3 OS=Porcine rotavirus A OX=10967 GN=NSP3 PE=3 SV=1
MM1 pKa = 7.81AEE3 pKa = 4.64LACFCYY9 pKa = 9.75PHH11 pKa = 7.6LEE13 pKa = 3.86NDD15 pKa = 3.51SYY17 pKa = 11.97KK18 pKa = 10.64FIPFNSLAIKK28 pKa = 10.62CMLTAKK34 pKa = 9.91VDD36 pKa = 3.91KK37 pKa = 10.63KK38 pKa = 11.12DD39 pKa = 3.04QDD41 pKa = 3.14KK42 pKa = 10.51FYY44 pKa = 11.42NSIIYY49 pKa = 9.99GIAPPPQFKK58 pKa = 10.27KK59 pKa = 10.52RR60 pKa = 11.84YY61 pKa = 5.79NTSDD65 pKa = 2.79NSRR68 pKa = 11.84GMNYY72 pKa = 7.22EE73 pKa = 3.56TTMFNKK79 pKa = 9.56VAALVCEE86 pKa = 4.32ALNSVKK92 pKa = 9.08ITQSDD97 pKa = 4.0LANVLSRR104 pKa = 11.84VVSVRR109 pKa = 11.84HH110 pKa = 5.93LEE112 pKa = 3.89NLVLRR117 pKa = 11.84KK118 pKa = 9.59EE119 pKa = 4.02NHH121 pKa = 5.63QDD123 pKa = 3.26VLFHH127 pKa = 6.93SKK129 pKa = 10.35EE130 pKa = 3.82LLLRR134 pKa = 11.84SVLIAIGQIKK144 pKa = 9.9EE145 pKa = 3.96IEE147 pKa = 4.25TTATAEE153 pKa = 3.87GGEE156 pKa = 4.08IVFQNAAFTMWKK168 pKa = 8.49LTYY171 pKa = 9.82LDD173 pKa = 4.31HH174 pKa = 8.04KK175 pKa = 10.88LMPILDD181 pKa = 3.78QNFIEE186 pKa = 4.65YY187 pKa = 10.13KK188 pKa = 9.01ITVNEE193 pKa = 4.63DD194 pKa = 2.89KK195 pKa = 10.59PISDD199 pKa = 3.42VHH201 pKa = 7.37IKK203 pKa = 10.54EE204 pKa = 4.36LVSEE208 pKa = 5.08LRR210 pKa = 11.84WQYY213 pKa = 11.88NKK215 pKa = 10.27FAVITHH221 pKa = 6.31GKK223 pKa = 7.43GHH225 pKa = 5.73YY226 pKa = 9.48RR227 pKa = 11.84VVKK230 pKa = 8.49YY231 pKa = 10.96SSVANHH237 pKa = 7.18ADD239 pKa = 3.3RR240 pKa = 11.84VFATFKK246 pKa = 11.24NNAKK250 pKa = 10.09SGNITEE256 pKa = 5.07FNLLDD261 pKa = 5.23QRR263 pKa = 11.84IIWQNWYY270 pKa = 10.72AFTSSMKK277 pKa = 10.28QGNTLDD283 pKa = 3.36VCKK286 pKa = 10.46RR287 pKa = 11.84LLFQKK292 pKa = 9.69MKK294 pKa = 10.49QEE296 pKa = 4.09KK297 pKa = 10.31NPFKK301 pKa = 10.91GLSTDD306 pKa = 2.99RR307 pKa = 11.84KK308 pKa = 8.31MDD310 pKa = 3.55EE311 pKa = 4.04VSQIGII317 pKa = 3.75
MM1 pKa = 7.81AEE3 pKa = 4.64LACFCYY9 pKa = 9.75PHH11 pKa = 7.6LEE13 pKa = 3.86NDD15 pKa = 3.51SYY17 pKa = 11.97KK18 pKa = 10.64FIPFNSLAIKK28 pKa = 10.62CMLTAKK34 pKa = 9.91VDD36 pKa = 3.91KK37 pKa = 10.63KK38 pKa = 11.12DD39 pKa = 3.04QDD41 pKa = 3.14KK42 pKa = 10.51FYY44 pKa = 11.42NSIIYY49 pKa = 9.99GIAPPPQFKK58 pKa = 10.27KK59 pKa = 10.52RR60 pKa = 11.84YY61 pKa = 5.79NTSDD65 pKa = 2.79NSRR68 pKa = 11.84GMNYY72 pKa = 7.22EE73 pKa = 3.56TTMFNKK79 pKa = 9.56VAALVCEE86 pKa = 4.32ALNSVKK92 pKa = 9.08ITQSDD97 pKa = 4.0LANVLSRR104 pKa = 11.84VVSVRR109 pKa = 11.84HH110 pKa = 5.93LEE112 pKa = 3.89NLVLRR117 pKa = 11.84KK118 pKa = 9.59EE119 pKa = 4.02NHH121 pKa = 5.63QDD123 pKa = 3.26VLFHH127 pKa = 6.93SKK129 pKa = 10.35EE130 pKa = 3.82LLLRR134 pKa = 11.84SVLIAIGQIKK144 pKa = 9.9EE145 pKa = 3.96IEE147 pKa = 4.25TTATAEE153 pKa = 3.87GGEE156 pKa = 4.08IVFQNAAFTMWKK168 pKa = 8.49LTYY171 pKa = 9.82LDD173 pKa = 4.31HH174 pKa = 8.04KK175 pKa = 10.88LMPILDD181 pKa = 3.78QNFIEE186 pKa = 4.65YY187 pKa = 10.13KK188 pKa = 9.01ITVNEE193 pKa = 4.63DD194 pKa = 2.89KK195 pKa = 10.59PISDD199 pKa = 3.42VHH201 pKa = 7.37IKK203 pKa = 10.54EE204 pKa = 4.36LVSEE208 pKa = 5.08LRR210 pKa = 11.84WQYY213 pKa = 11.88NKK215 pKa = 10.27FAVITHH221 pKa = 6.31GKK223 pKa = 7.43GHH225 pKa = 5.73YY226 pKa = 9.48RR227 pKa = 11.84VVKK230 pKa = 8.49YY231 pKa = 10.96SSVANHH237 pKa = 7.18ADD239 pKa = 3.3RR240 pKa = 11.84VFATFKK246 pKa = 11.24NNAKK250 pKa = 10.09SGNITEE256 pKa = 5.07FNLLDD261 pKa = 5.23QRR263 pKa = 11.84IIWQNWYY270 pKa = 10.72AFTSSMKK277 pKa = 10.28QGNTLDD283 pKa = 3.36VCKK286 pKa = 10.46RR287 pKa = 11.84LLFQKK292 pKa = 9.69MKK294 pKa = 10.49QEE296 pKa = 4.09KK297 pKa = 10.31NPFKK301 pKa = 10.91GLSTDD306 pKa = 2.99RR307 pKa = 11.84KK308 pKa = 8.31MDD310 pKa = 3.55EE311 pKa = 4.04VSQIGII317 pKa = 3.75
Molecular weight: 36.58 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3797 |
175 |
517 |
345.2 |
39.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.083 ± 0.379 | 1.422 ± 0.359 |
6.321 ± 0.439 | 5.952 ± 0.572 |
4.135 ± 0.33 | 3.081 ± 0.267 |
1.844 ± 0.331 | 7.453 ± 0.315 |
6.769 ± 0.606 | 9.033 ± 0.383 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.213 ± 0.288 | 6.821 ± 0.473 |
3.424 ± 0.317 | 4.214 ± 0.433 |
4.767 ± 0.326 | 7.585 ± 0.798 |
6.61 ± 0.633 | 6.11 ± 0.272 |
1.317 ± 0.17 | 4.846 ± 0.483 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
