
Aggregicoccus sp. 17bor-14
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Myxococcales; Cystobacterineae; Myxococcaceae; Aggregicoccus; unclassified Aggregicoccus
Average proteome isoelectric point is 7.04
Get precalculated fractions of proteins
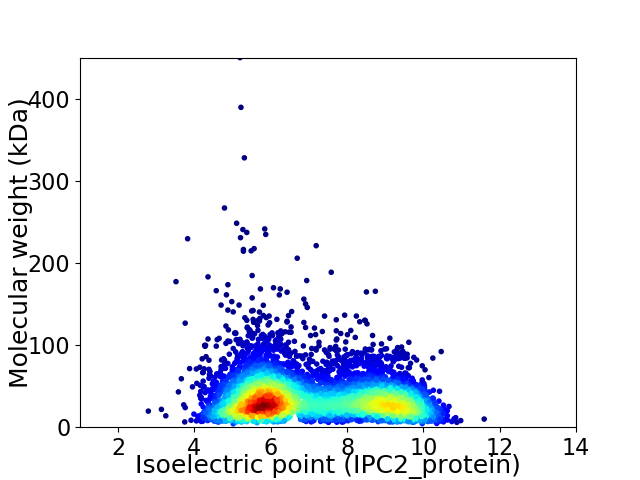
Virtual 2D-PAGE plot for 5829 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6I2GT76|A0A6I2GT76_9DELT GGDEF domain-containing protein OS=Aggregicoccus sp. 17bor-14 OX=2583808 GN=FGE12_25110 PE=4 SV=1
MM1 pKa = 7.7RR2 pKa = 11.84AALLCAGLLALAGCGGSPDD21 pKa = 4.14PEE23 pKa = 4.19PVKK26 pKa = 9.29PTPPVTPAPPVVVDD40 pKa = 4.28ADD42 pKa = 3.78HH43 pKa = 7.51DD44 pKa = 4.61GVPSTADD51 pKa = 3.65CADD54 pKa = 4.19DD55 pKa = 4.1DD56 pKa = 4.12ATRR59 pKa = 11.84FQYY62 pKa = 11.15VSGHH66 pKa = 6.47RR67 pKa = 11.84DD68 pKa = 3.01ADD70 pKa = 3.85GDD72 pKa = 4.42GVGADD77 pKa = 3.91ALEE80 pKa = 4.23QVCAGAALPQGWVSTGGDD98 pKa = 3.74CATYY102 pKa = 10.7DD103 pKa = 3.31ATRR106 pKa = 11.84WRR108 pKa = 11.84EE109 pKa = 3.67LAVYY113 pKa = 9.49EE114 pKa = 4.6DD115 pKa = 3.76WDD117 pKa = 3.76GDD119 pKa = 3.57GRR121 pKa = 11.84TRR123 pKa = 11.84PYY125 pKa = 11.19AQTLCIGAQVPTGYY139 pKa = 8.47VTQRR143 pKa = 11.84GEE145 pKa = 4.15DD146 pKa = 3.53DD147 pKa = 4.94CSDD150 pKa = 3.73FDD152 pKa = 3.38ATAWHH157 pKa = 6.27EE158 pKa = 4.24VPLYY162 pKa = 10.7FDD164 pKa = 4.91LDD166 pKa = 3.75GDD168 pKa = 4.76GVGDD172 pKa = 4.29DD173 pKa = 4.64YY174 pKa = 12.1AMSMCLGSAPPPTYY188 pKa = 9.96MVATGGDD195 pKa = 3.97CAPRR199 pKa = 11.84DD200 pKa = 3.61ATLYY204 pKa = 10.58TMLPYY209 pKa = 10.3AYY211 pKa = 9.4RR212 pKa = 11.84DD213 pKa = 3.47ADD215 pKa = 3.64GDD217 pKa = 4.16GATVPQQGSVCSGFYY232 pKa = 10.76LPAGYY237 pKa = 9.86RR238 pKa = 11.84EE239 pKa = 4.22SAQGLDD245 pKa = 4.71CNDD248 pKa = 4.39ADD250 pKa = 3.86PSVYY254 pKa = 10.76SMQPGFPDD262 pKa = 3.85PDD264 pKa = 4.08GDD266 pKa = 4.2GVGSGEE272 pKa = 4.21SFEE275 pKa = 4.53VCAGVAMPRR284 pKa = 11.84YY285 pKa = 9.17SSRR288 pKa = 11.84RR289 pKa = 11.84SDD291 pKa = 3.32DD292 pKa = 3.78CAPQDD297 pKa = 3.5SSRR300 pKa = 11.84WEE302 pKa = 3.64QRR304 pKa = 11.84EE305 pKa = 3.82YY306 pKa = 11.38RR307 pKa = 11.84LGDD310 pKa = 3.36ADD312 pKa = 3.82GDD314 pKa = 4.01GRR316 pKa = 11.84TVPLAEE322 pKa = 4.28PASFCVGNTDD332 pKa = 3.18PQGYY336 pKa = 9.63SRR338 pKa = 11.84GTPWPDD344 pKa = 3.35DD345 pKa = 4.19CDD347 pKa = 4.01DD348 pKa = 4.42ADD350 pKa = 3.54AARR353 pKa = 11.84YY354 pKa = 8.42QVLAYY359 pKa = 10.09AYY361 pKa = 10.04RR362 pKa = 11.84DD363 pKa = 3.5ADD365 pKa = 3.59GDD367 pKa = 4.21GATVPATGSLCSGASLPAGYY387 pKa = 8.17ATQSRR392 pKa = 11.84GADD395 pKa = 3.64CDD397 pKa = 4.08DD398 pKa = 4.7ADD400 pKa = 3.72AQRR403 pKa = 11.84FVQLSGFADD412 pKa = 3.39VDD414 pKa = 3.72ADD416 pKa = 3.94GVGAGEE422 pKa = 4.03AQAFCTAGALPAGFVASSTDD442 pKa = 3.62CAAQDD447 pKa = 3.38AARR450 pKa = 11.84WRR452 pKa = 11.84TVTPGFLDD460 pKa = 3.28QDD462 pKa = 3.56GDD464 pKa = 4.11GYY466 pKa = 9.48TVVDD470 pKa = 4.41PAPTAQCIGTAPEE483 pKa = 4.32APSVLAARR491 pKa = 11.84GNDD494 pKa = 4.27CEE496 pKa = 4.7DD497 pKa = 3.51TDD499 pKa = 3.74PTRR502 pKa = 11.84FLWRR506 pKa = 11.84VFYY509 pKa = 10.68RR510 pKa = 11.84DD511 pKa = 3.2EE512 pKa = 6.4DD513 pKa = 4.37GDD515 pKa = 4.45GVGAAPRR522 pKa = 11.84LLRR525 pKa = 11.84CLATGAAPAGEE536 pKa = 4.7SPYY539 pKa = 11.33GWDD542 pKa = 5.03SDD544 pKa = 4.27DD545 pKa = 5.07ADD547 pKa = 3.94PAVQQSEE554 pKa = 4.07EE555 pKa = 4.3DD556 pKa = 3.73EE557 pKa = 5.08AVLQLLLEE565 pKa = 4.42TT566 pKa = 4.86
MM1 pKa = 7.7RR2 pKa = 11.84AALLCAGLLALAGCGGSPDD21 pKa = 4.14PEE23 pKa = 4.19PVKK26 pKa = 9.29PTPPVTPAPPVVVDD40 pKa = 4.28ADD42 pKa = 3.78HH43 pKa = 7.51DD44 pKa = 4.61GVPSTADD51 pKa = 3.65CADD54 pKa = 4.19DD55 pKa = 4.1DD56 pKa = 4.12ATRR59 pKa = 11.84FQYY62 pKa = 11.15VSGHH66 pKa = 6.47RR67 pKa = 11.84DD68 pKa = 3.01ADD70 pKa = 3.85GDD72 pKa = 4.42GVGADD77 pKa = 3.91ALEE80 pKa = 4.23QVCAGAALPQGWVSTGGDD98 pKa = 3.74CATYY102 pKa = 10.7DD103 pKa = 3.31ATRR106 pKa = 11.84WRR108 pKa = 11.84EE109 pKa = 3.67LAVYY113 pKa = 9.49EE114 pKa = 4.6DD115 pKa = 3.76WDD117 pKa = 3.76GDD119 pKa = 3.57GRR121 pKa = 11.84TRR123 pKa = 11.84PYY125 pKa = 11.19AQTLCIGAQVPTGYY139 pKa = 8.47VTQRR143 pKa = 11.84GEE145 pKa = 4.15DD146 pKa = 3.53DD147 pKa = 4.94CSDD150 pKa = 3.73FDD152 pKa = 3.38ATAWHH157 pKa = 6.27EE158 pKa = 4.24VPLYY162 pKa = 10.7FDD164 pKa = 4.91LDD166 pKa = 3.75GDD168 pKa = 4.76GVGDD172 pKa = 4.29DD173 pKa = 4.64YY174 pKa = 12.1AMSMCLGSAPPPTYY188 pKa = 9.96MVATGGDD195 pKa = 3.97CAPRR199 pKa = 11.84DD200 pKa = 3.61ATLYY204 pKa = 10.58TMLPYY209 pKa = 10.3AYY211 pKa = 9.4RR212 pKa = 11.84DD213 pKa = 3.47ADD215 pKa = 3.64GDD217 pKa = 4.16GATVPQQGSVCSGFYY232 pKa = 10.76LPAGYY237 pKa = 9.86RR238 pKa = 11.84EE239 pKa = 4.22SAQGLDD245 pKa = 4.71CNDD248 pKa = 4.39ADD250 pKa = 3.86PSVYY254 pKa = 10.76SMQPGFPDD262 pKa = 3.85PDD264 pKa = 4.08GDD266 pKa = 4.2GVGSGEE272 pKa = 4.21SFEE275 pKa = 4.53VCAGVAMPRR284 pKa = 11.84YY285 pKa = 9.17SSRR288 pKa = 11.84RR289 pKa = 11.84SDD291 pKa = 3.32DD292 pKa = 3.78CAPQDD297 pKa = 3.5SSRR300 pKa = 11.84WEE302 pKa = 3.64QRR304 pKa = 11.84EE305 pKa = 3.82YY306 pKa = 11.38RR307 pKa = 11.84LGDD310 pKa = 3.36ADD312 pKa = 3.82GDD314 pKa = 4.01GRR316 pKa = 11.84TVPLAEE322 pKa = 4.28PASFCVGNTDD332 pKa = 3.18PQGYY336 pKa = 9.63SRR338 pKa = 11.84GTPWPDD344 pKa = 3.35DD345 pKa = 4.19CDD347 pKa = 4.01DD348 pKa = 4.42ADD350 pKa = 3.54AARR353 pKa = 11.84YY354 pKa = 8.42QVLAYY359 pKa = 10.09AYY361 pKa = 10.04RR362 pKa = 11.84DD363 pKa = 3.5ADD365 pKa = 3.59GDD367 pKa = 4.21GATVPATGSLCSGASLPAGYY387 pKa = 8.17ATQSRR392 pKa = 11.84GADD395 pKa = 3.64CDD397 pKa = 4.08DD398 pKa = 4.7ADD400 pKa = 3.72AQRR403 pKa = 11.84FVQLSGFADD412 pKa = 3.39VDD414 pKa = 3.72ADD416 pKa = 3.94GVGAGEE422 pKa = 4.03AQAFCTAGALPAGFVASSTDD442 pKa = 3.62CAAQDD447 pKa = 3.38AARR450 pKa = 11.84WRR452 pKa = 11.84TVTPGFLDD460 pKa = 3.28QDD462 pKa = 3.56GDD464 pKa = 4.11GYY466 pKa = 9.48TVVDD470 pKa = 4.41PAPTAQCIGTAPEE483 pKa = 4.32APSVLAARR491 pKa = 11.84GNDD494 pKa = 4.27CEE496 pKa = 4.7DD497 pKa = 3.51TDD499 pKa = 3.74PTRR502 pKa = 11.84FLWRR506 pKa = 11.84VFYY509 pKa = 10.68RR510 pKa = 11.84DD511 pKa = 3.2EE512 pKa = 6.4DD513 pKa = 4.37GDD515 pKa = 4.45GVGAAPRR522 pKa = 11.84LLRR525 pKa = 11.84CLATGAAPAGEE536 pKa = 4.7SPYY539 pKa = 11.33GWDD542 pKa = 5.03SDD544 pKa = 4.27DD545 pKa = 5.07ADD547 pKa = 3.94PAVQQSEE554 pKa = 4.07EE555 pKa = 4.3DD556 pKa = 3.73EE557 pKa = 5.08AVLQLLLEE565 pKa = 4.42TT566 pKa = 4.86
Molecular weight: 58.97 kDa
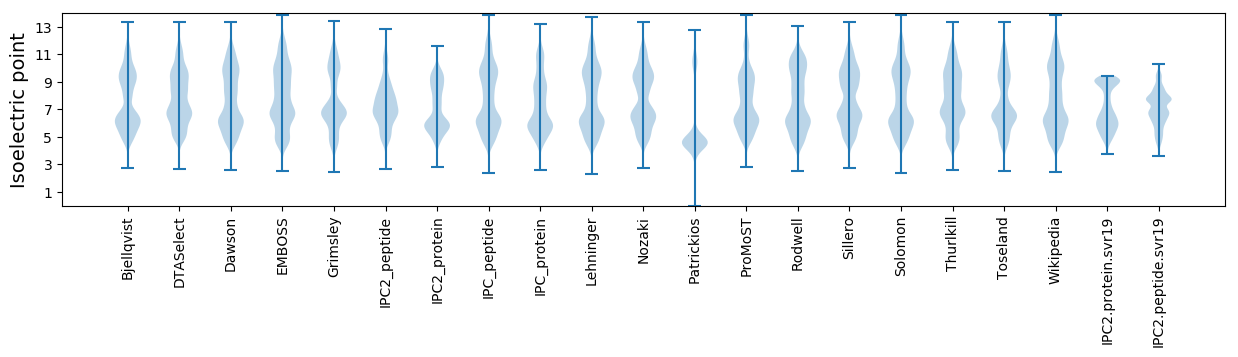
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6I2GLG1|A0A6I2GLG1_9DELT TIGR02266 family protein OS=Aggregicoccus sp. 17bor-14 OX=2583808 GN=FGE12_00710 PE=4 SV=1
MM1 pKa = 7.76AARR4 pKa = 11.84KK5 pKa = 7.51TSRR8 pKa = 11.84KK9 pKa = 8.11STAKK13 pKa = 10.09RR14 pKa = 11.84STGTRR19 pKa = 11.84SKK21 pKa = 10.82RR22 pKa = 11.84SGGAKK27 pKa = 9.55RR28 pKa = 11.84RR29 pKa = 11.84TGGTAAKK36 pKa = 9.97RR37 pKa = 11.84ATAKK41 pKa = 10.47RR42 pKa = 11.84STAKK46 pKa = 10.4RR47 pKa = 11.84GTARR51 pKa = 11.84KK52 pKa = 9.0SASRR56 pKa = 11.84GKK58 pKa = 8.93RR59 pKa = 11.84TAKK62 pKa = 10.2RR63 pKa = 11.84STAGKK68 pKa = 9.63RR69 pKa = 11.84VTARR73 pKa = 11.84KK74 pKa = 8.82PGARR78 pKa = 11.84RR79 pKa = 11.84TAAKK83 pKa = 8.34RR84 pKa = 11.84TSTRR88 pKa = 11.84RR89 pKa = 11.84ARR91 pKa = 11.84RR92 pKa = 3.31
MM1 pKa = 7.76AARR4 pKa = 11.84KK5 pKa = 7.51TSRR8 pKa = 11.84KK9 pKa = 8.11STAKK13 pKa = 10.09RR14 pKa = 11.84STGTRR19 pKa = 11.84SKK21 pKa = 10.82RR22 pKa = 11.84SGGAKK27 pKa = 9.55RR28 pKa = 11.84RR29 pKa = 11.84TGGTAAKK36 pKa = 9.97RR37 pKa = 11.84ATAKK41 pKa = 10.47RR42 pKa = 11.84STAKK46 pKa = 10.4RR47 pKa = 11.84GTARR51 pKa = 11.84KK52 pKa = 9.0SASRR56 pKa = 11.84GKK58 pKa = 8.93RR59 pKa = 11.84TAKK62 pKa = 10.2RR63 pKa = 11.84STAGKK68 pKa = 9.63RR69 pKa = 11.84VTARR73 pKa = 11.84KK74 pKa = 8.82PGARR78 pKa = 11.84RR79 pKa = 11.84TAAKK83 pKa = 8.34RR84 pKa = 11.84TSTRR88 pKa = 11.84RR89 pKa = 11.84ARR91 pKa = 11.84RR92 pKa = 3.31
Molecular weight: 9.91 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2075293 |
38 |
4099 |
356.0 |
38.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.817 ± 0.055 | 0.871 ± 0.015 |
4.643 ± 0.023 | 6.348 ± 0.038 |
3.174 ± 0.02 | 9.064 ± 0.033 |
2.082 ± 0.016 | 2.416 ± 0.027 |
2.594 ± 0.031 | 11.991 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.645 ± 0.013 | 1.798 ± 0.019 |
6.227 ± 0.033 | 3.591 ± 0.021 |
8.38 ± 0.041 | 5.215 ± 0.025 |
4.85 ± 0.028 | 8.001 ± 0.028 |
1.243 ± 0.015 | 2.049 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
