
Histidinibacterium lentulum
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Histidinibacterium
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins
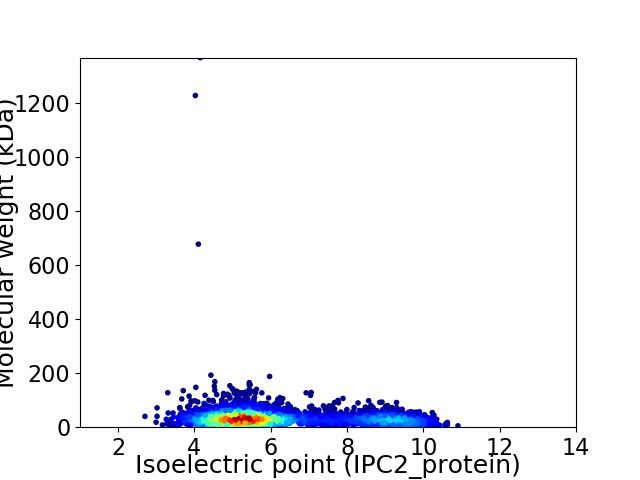
Virtual 2D-PAGE plot for 4017 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3N2R8B2|A0A3N2R8B2_9RHOB ABC transporter permease OS=Histidinibacterium lentulum OX=2480588 GN=EAT49_05195 PE=4 SV=1
MM1 pKa = 6.72KK2 pKa = 9.93TSVSILAMAAATIGAPHH19 pKa = 6.78FAAAQDD25 pKa = 3.77APTIATVVKK34 pKa = 10.13IAGIQWFNRR43 pKa = 11.84MEE45 pKa = 4.29EE46 pKa = 4.29GVNQYY51 pKa = 11.1AADD54 pKa = 3.61TGANAFQVGPAQADD68 pKa = 3.76PQQQAALIEE77 pKa = 4.32DD78 pKa = 4.95MIAQGVDD85 pKa = 2.94ALAVVPMSPEE95 pKa = 3.51ALEE98 pKa = 4.31PVLGRR103 pKa = 11.84AMEE106 pKa = 3.92QGIVVITHH114 pKa = 6.45EE115 pKa = 4.85AASQQNTMYY124 pKa = 10.68DD125 pKa = 2.64IEE127 pKa = 4.88AFVNEE132 pKa = 4.48DD133 pKa = 3.2FGANLMEE140 pKa = 5.55QMAQCMGGEE149 pKa = 4.23GEE151 pKa = 4.26YY152 pKa = 10.91AVFVGSLTSQTHH164 pKa = 4.79NQWVDD169 pKa = 2.95GAIAHH174 pKa = 6.2QEE176 pKa = 3.63ANYY179 pKa = 10.48PNMTLVGDD187 pKa = 4.0KK188 pKa = 11.38NEE190 pKa = 4.42TFDD193 pKa = 5.46DD194 pKa = 4.03AQQAYY199 pKa = 8.66EE200 pKa = 3.91AAQEE204 pKa = 4.15VMRR207 pKa = 11.84AFPNVRR213 pKa = 11.84GMQGSASTDD222 pKa = 3.2VAGIGLAVEE231 pKa = 4.06EE232 pKa = 4.93RR233 pKa = 11.84GMEE236 pKa = 4.23DD237 pKa = 3.15ATCVFGTSLPSIAGQYY253 pKa = 10.54LEE255 pKa = 4.52TGAVDD260 pKa = 6.37GIGFWDD266 pKa = 4.36PSVAGYY272 pKa = 11.49AMNKK276 pKa = 7.23VAQMVMNGEE285 pKa = 4.26EE286 pKa = 4.33VTDD289 pKa = 4.41GMDD292 pKa = 3.86LGLEE296 pKa = 4.41GYY298 pKa = 10.7EE299 pKa = 4.53NVTLDD304 pKa = 3.28GKK306 pKa = 10.65VIYY309 pKa = 9.58GQAWVNVNADD319 pKa = 3.13NMADD323 pKa = 3.64YY324 pKa = 10.2PFF326 pKa = 5.35
MM1 pKa = 6.72KK2 pKa = 9.93TSVSILAMAAATIGAPHH19 pKa = 6.78FAAAQDD25 pKa = 3.77APTIATVVKK34 pKa = 10.13IAGIQWFNRR43 pKa = 11.84MEE45 pKa = 4.29EE46 pKa = 4.29GVNQYY51 pKa = 11.1AADD54 pKa = 3.61TGANAFQVGPAQADD68 pKa = 3.76PQQQAALIEE77 pKa = 4.32DD78 pKa = 4.95MIAQGVDD85 pKa = 2.94ALAVVPMSPEE95 pKa = 3.51ALEE98 pKa = 4.31PVLGRR103 pKa = 11.84AMEE106 pKa = 3.92QGIVVITHH114 pKa = 6.45EE115 pKa = 4.85AASQQNTMYY124 pKa = 10.68DD125 pKa = 2.64IEE127 pKa = 4.88AFVNEE132 pKa = 4.48DD133 pKa = 3.2FGANLMEE140 pKa = 5.55QMAQCMGGEE149 pKa = 4.23GEE151 pKa = 4.26YY152 pKa = 10.91AVFVGSLTSQTHH164 pKa = 4.79NQWVDD169 pKa = 2.95GAIAHH174 pKa = 6.2QEE176 pKa = 3.63ANYY179 pKa = 10.48PNMTLVGDD187 pKa = 4.0KK188 pKa = 11.38NEE190 pKa = 4.42TFDD193 pKa = 5.46DD194 pKa = 4.03AQQAYY199 pKa = 8.66EE200 pKa = 3.91AAQEE204 pKa = 4.15VMRR207 pKa = 11.84AFPNVRR213 pKa = 11.84GMQGSASTDD222 pKa = 3.2VAGIGLAVEE231 pKa = 4.06EE232 pKa = 4.93RR233 pKa = 11.84GMEE236 pKa = 4.23DD237 pKa = 3.15ATCVFGTSLPSIAGQYY253 pKa = 10.54LEE255 pKa = 4.52TGAVDD260 pKa = 6.37GIGFWDD266 pKa = 4.36PSVAGYY272 pKa = 11.49AMNKK276 pKa = 7.23VAQMVMNGEE285 pKa = 4.26EE286 pKa = 4.33VTDD289 pKa = 4.41GMDD292 pKa = 3.86LGLEE296 pKa = 4.41GYY298 pKa = 10.7EE299 pKa = 4.53NVTLDD304 pKa = 3.28GKK306 pKa = 10.65VIYY309 pKa = 9.58GQAWVNVNADD319 pKa = 3.13NMADD323 pKa = 3.64YY324 pKa = 10.2PFF326 pKa = 5.35
Molecular weight: 34.58 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3N2R5D8|A0A3N2R5D8_9RHOB NAD-dependent epimerase/dehydratase family protein OS=Histidinibacterium lentulum OX=2480588 GN=EAT49_10345 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84VRR12 pKa = 11.84KK13 pKa = 8.99NRR15 pKa = 11.84HH16 pKa = 3.77GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.43GGRR28 pKa = 11.84KK29 pKa = 8.99VLNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.57SLCAA44 pKa = 3.82
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84VRR12 pKa = 11.84KK13 pKa = 8.99NRR15 pKa = 11.84HH16 pKa = 3.77GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.43GGRR28 pKa = 11.84KK29 pKa = 8.99VLNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.57SLCAA44 pKa = 3.82
Molecular weight: 5.13 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1290838 |
28 |
12941 |
321.3 |
34.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.439 ± 0.065 | 0.811 ± 0.016 |
5.99 ± 0.056 | 6.248 ± 0.04 |
3.486 ± 0.028 | 9.332 ± 0.067 |
1.956 ± 0.023 | 4.579 ± 0.031 |
2.095 ± 0.034 | 10.3 ± 0.066 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.504 ± 0.031 | 2.053 ± 0.028 |
5.558 ± 0.045 | 2.617 ± 0.02 |
7.814 ± 0.061 | 4.745 ± 0.042 |
5.481 ± 0.062 | 7.513 ± 0.043 |
1.47 ± 0.022 | 2.008 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
