
Mastomys natalensis papillomavirus (isolate African multimammate rat) (MnPV)
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Iotapapillomavirus; Iotapapillomavirus 1
Average proteome isoelectric point is 6.81
Get precalculated fractions of proteins
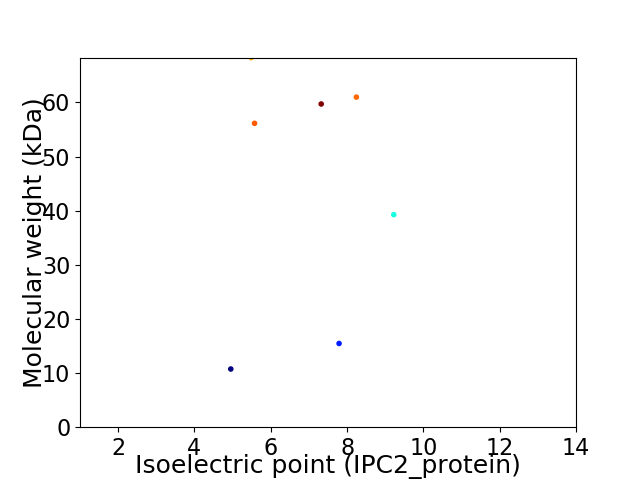
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q84356|VE1_MNPVA Replication protein E1 OS=Mastomys natalensis papillomavirus (isolate African multimammate rat) OX=654915 GN=E1 PE=3 SV=1
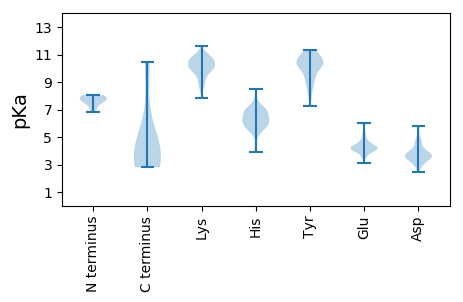
MM1 pKa = 7.73IGPDD5 pKa = 3.27TTRR8 pKa = 11.84CLTGEE13 pKa = 4.27TPDD16 pKa = 3.94SVSLYY21 pKa = 9.25CHH23 pKa = 6.29EE24 pKa = 4.63VLDD27 pKa = 4.21EE28 pKa = 5.19DD29 pKa = 4.24EE30 pKa = 5.22LKK32 pKa = 10.87EE33 pKa = 4.13PTEE36 pKa = 4.04AAPPPEE42 pKa = 4.54QYY44 pKa = 10.07TLYY47 pKa = 10.4QVLIEE52 pKa = 4.64CPEE55 pKa = 4.09CNKK58 pKa = 10.01TIRR61 pKa = 11.84LTCAAQAHH69 pKa = 5.57QIRR72 pKa = 11.84GLEE75 pKa = 3.87HH76 pKa = 7.04LLLDD80 pKa = 4.47GLRR83 pKa = 11.84VICPRR88 pKa = 11.84CNQKK92 pKa = 10.3NGRR95 pKa = 11.84SS96 pKa = 3.52
MM1 pKa = 7.73IGPDD5 pKa = 3.27TTRR8 pKa = 11.84CLTGEE13 pKa = 4.27TPDD16 pKa = 3.94SVSLYY21 pKa = 9.25CHH23 pKa = 6.29EE24 pKa = 4.63VLDD27 pKa = 4.21EE28 pKa = 5.19DD29 pKa = 4.24EE30 pKa = 5.22LKK32 pKa = 10.87EE33 pKa = 4.13PTEE36 pKa = 4.04AAPPPEE42 pKa = 4.54QYY44 pKa = 10.07TLYY47 pKa = 10.4QVLIEE52 pKa = 4.64CPEE55 pKa = 4.09CNKK58 pKa = 10.01TIRR61 pKa = 11.84LTCAAQAHH69 pKa = 5.57QIRR72 pKa = 11.84GLEE75 pKa = 3.87HH76 pKa = 7.04LLLDD80 pKa = 4.47GLRR83 pKa = 11.84VICPRR88 pKa = 11.84CNQKK92 pKa = 10.3NGRR95 pKa = 11.84SS96 pKa = 3.52
Molecular weight: 10.75 kDa
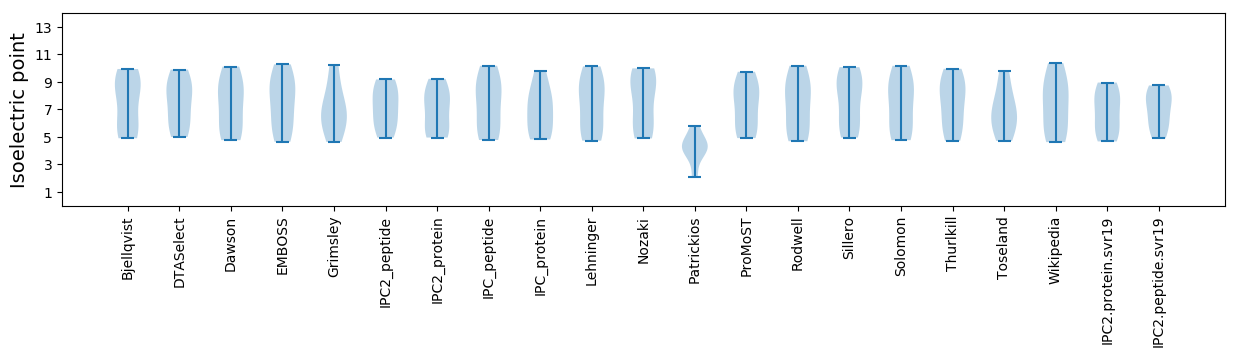
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P30735|VE6_MNPVA Protein E6 OS=Mastomys natalensis papillomavirus (isolate African multimammate rat) OX=654915 GN=E6 PE=3 SV=2
MM1 pKa = 8.05RR2 pKa = 11.84LQILLRR8 pKa = 11.84GFSSTPQTGFPRR20 pKa = 11.84GDD22 pKa = 3.42PVRR25 pKa = 11.84LHH27 pKa = 6.82GNTTTGLPIPLRR39 pKa = 11.84NSSSNQILLRR49 pKa = 11.84EE50 pKa = 4.23GRR52 pKa = 11.84GDD54 pKa = 3.69YY55 pKa = 10.53PDD57 pKa = 3.53GARR60 pKa = 11.84RR61 pKa = 11.84EE62 pKa = 4.06TRR64 pKa = 11.84RR65 pKa = 11.84YY66 pKa = 8.37YY67 pKa = 10.3QGPTPTPRR75 pKa = 11.84SLSPPIYY82 pKa = 9.83RR83 pKa = 11.84PPPSYY88 pKa = 10.45EE89 pKa = 3.37EE90 pKa = 3.64SRR92 pKa = 11.84RR93 pKa = 11.84RR94 pKa = 11.84RR95 pKa = 11.84KK96 pKa = 9.53LRR98 pKa = 11.84RR99 pKa = 11.84RR100 pKa = 11.84QDD102 pKa = 3.01GRR104 pKa = 11.84VKK106 pKa = 10.87YY107 pKa = 9.06PASPYY112 pKa = 8.08RR113 pKa = 11.84TKK115 pKa = 10.91PPGEE119 pKa = 4.08TSSDD123 pKa = 3.7DD124 pKa = 3.57EE125 pKa = 4.89DD126 pKa = 3.78EE127 pKa = 4.47GRR129 pKa = 11.84GGHH132 pKa = 5.71EE133 pKa = 3.86PRR135 pKa = 11.84PQRR138 pKa = 11.84RR139 pKa = 11.84LPRR142 pKa = 11.84GLRR145 pKa = 11.84DD146 pKa = 3.27RR147 pKa = 11.84GEE149 pKa = 4.09RR150 pKa = 11.84APEE153 pKa = 3.63RR154 pKa = 11.84RR155 pKa = 11.84RR156 pKa = 11.84PPVQEE161 pKa = 4.08GEE163 pKa = 4.05EE164 pKa = 4.18DD165 pKa = 3.49VDD167 pKa = 4.63GVGALLDD174 pKa = 4.04DD175 pKa = 4.61LKK177 pKa = 11.15LYY179 pKa = 10.23QEE181 pKa = 4.64PPGDD185 pKa = 3.74PVEE188 pKa = 5.78DD189 pKa = 3.64SDD191 pKa = 5.49SPGSRR196 pKa = 11.84LTPAPPDD203 pKa = 3.4LSRR206 pKa = 11.84YY207 pKa = 10.24DD208 pKa = 3.61STRR211 pKa = 11.84LQVDD215 pKa = 4.07AEE217 pKa = 4.35SSPPRR222 pKa = 11.84TPRR225 pKa = 11.84PAPTLVAEE233 pKa = 4.61CTPGRR238 pKa = 11.84PSPQTGSGQQALGEE252 pKa = 4.28PPSRR256 pKa = 11.84PSRR259 pKa = 11.84GHH261 pKa = 6.15CRR263 pKa = 11.84DD264 pKa = 3.19PRR266 pKa = 11.84TACLLIIKK274 pKa = 9.99GSSNQVKK281 pKa = 10.18CLRR284 pKa = 11.84FRR286 pKa = 11.84LKK288 pKa = 10.12SWHH291 pKa = 6.87HH292 pKa = 6.06SLFSYY297 pKa = 10.66ISTTWQWVPSVGSNRR312 pKa = 11.84IGRR315 pKa = 11.84SRR317 pKa = 11.84ILVMCEE323 pKa = 4.92DD324 pKa = 4.12SAQMDD329 pKa = 3.63RR330 pKa = 11.84FLCTVKK336 pKa = 10.23IPAGMTVEE344 pKa = 4.22QCSMASVV351 pKa = 3.38
MM1 pKa = 8.05RR2 pKa = 11.84LQILLRR8 pKa = 11.84GFSSTPQTGFPRR20 pKa = 11.84GDD22 pKa = 3.42PVRR25 pKa = 11.84LHH27 pKa = 6.82GNTTTGLPIPLRR39 pKa = 11.84NSSSNQILLRR49 pKa = 11.84EE50 pKa = 4.23GRR52 pKa = 11.84GDD54 pKa = 3.69YY55 pKa = 10.53PDD57 pKa = 3.53GARR60 pKa = 11.84RR61 pKa = 11.84EE62 pKa = 4.06TRR64 pKa = 11.84RR65 pKa = 11.84YY66 pKa = 8.37YY67 pKa = 10.3QGPTPTPRR75 pKa = 11.84SLSPPIYY82 pKa = 9.83RR83 pKa = 11.84PPPSYY88 pKa = 10.45EE89 pKa = 3.37EE90 pKa = 3.64SRR92 pKa = 11.84RR93 pKa = 11.84RR94 pKa = 11.84RR95 pKa = 11.84KK96 pKa = 9.53LRR98 pKa = 11.84RR99 pKa = 11.84RR100 pKa = 11.84QDD102 pKa = 3.01GRR104 pKa = 11.84VKK106 pKa = 10.87YY107 pKa = 9.06PASPYY112 pKa = 8.08RR113 pKa = 11.84TKK115 pKa = 10.91PPGEE119 pKa = 4.08TSSDD123 pKa = 3.7DD124 pKa = 3.57EE125 pKa = 4.89DD126 pKa = 3.78EE127 pKa = 4.47GRR129 pKa = 11.84GGHH132 pKa = 5.71EE133 pKa = 3.86PRR135 pKa = 11.84PQRR138 pKa = 11.84RR139 pKa = 11.84LPRR142 pKa = 11.84GLRR145 pKa = 11.84DD146 pKa = 3.27RR147 pKa = 11.84GEE149 pKa = 4.09RR150 pKa = 11.84APEE153 pKa = 3.63RR154 pKa = 11.84RR155 pKa = 11.84RR156 pKa = 11.84PPVQEE161 pKa = 4.08GEE163 pKa = 4.05EE164 pKa = 4.18DD165 pKa = 3.49VDD167 pKa = 4.63GVGALLDD174 pKa = 4.04DD175 pKa = 4.61LKK177 pKa = 11.15LYY179 pKa = 10.23QEE181 pKa = 4.64PPGDD185 pKa = 3.74PVEE188 pKa = 5.78DD189 pKa = 3.64SDD191 pKa = 5.49SPGSRR196 pKa = 11.84LTPAPPDD203 pKa = 3.4LSRR206 pKa = 11.84YY207 pKa = 10.24DD208 pKa = 3.61STRR211 pKa = 11.84LQVDD215 pKa = 4.07AEE217 pKa = 4.35SSPPRR222 pKa = 11.84TPRR225 pKa = 11.84PAPTLVAEE233 pKa = 4.61CTPGRR238 pKa = 11.84PSPQTGSGQQALGEE252 pKa = 4.28PPSRR256 pKa = 11.84PSRR259 pKa = 11.84GHH261 pKa = 6.15CRR263 pKa = 11.84DD264 pKa = 3.19PRR266 pKa = 11.84TACLLIIKK274 pKa = 9.99GSSNQVKK281 pKa = 10.18CLRR284 pKa = 11.84FRR286 pKa = 11.84LKK288 pKa = 10.12SWHH291 pKa = 6.87HH292 pKa = 6.06SLFSYY297 pKa = 10.66ISTTWQWVPSVGSNRR312 pKa = 11.84IGRR315 pKa = 11.84SRR317 pKa = 11.84ILVMCEE323 pKa = 4.92DD324 pKa = 4.12SAQMDD329 pKa = 3.63RR330 pKa = 11.84FLCTVKK336 pKa = 10.23IPAGMTVEE344 pKa = 4.22QCSMASVV351 pKa = 3.38
Molecular weight: 39.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2777 |
96 |
602 |
396.7 |
44.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.834 ± 0.503 | 2.269 ± 0.572 |
5.654 ± 0.142 | 6.41 ± 0.532 |
2.953 ± 0.512 | 7.562 ± 0.674 |
2.233 ± 0.172 | 4.105 ± 0.463 |
3.925 ± 0.624 | 7.922 ± 0.63 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.125 ± 0.295 | 3.241 ± 0.677 |
7.814 ± 1.118 | 3.673 ± 0.422 |
8.39 ± 1.337 | 8.57 ± 0.696 |
6.914 ± 0.371 | 5.402 ± 0.378 |
1.404 ± 0.223 | 3.601 ± 0.441 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
