
Physostegia chlorotic mottle virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Betarhabdovirinae; Alphanucleorhabdovirus; Physostegia chlorotic mottle alphanucleorhabdovirus
Average proteome isoelectric point is 6.06
Get precalculated fractions of proteins
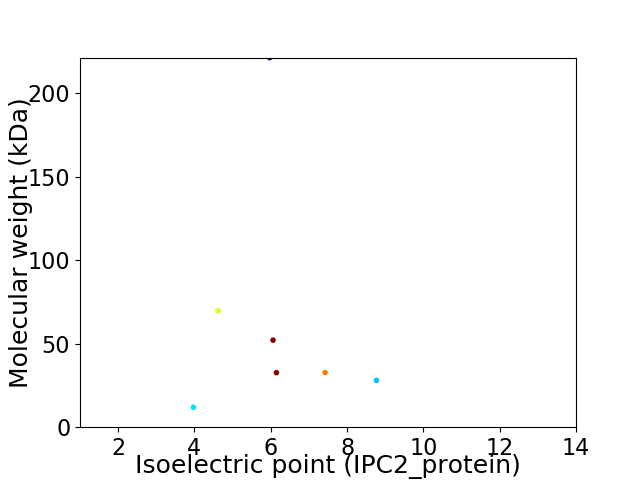
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1D8FVH3|A0A1D8FVH3_9RHAB Putative movement protein OS=Physostegia chlorotic mottle virus OX=1905830 GN=Y PE=4 SV=1
MM1 pKa = 7.18SASRR5 pKa = 11.84NTTTAPDD12 pKa = 3.52KK13 pKa = 10.79PGNQGDD19 pKa = 3.91EE20 pKa = 4.45IFIEE24 pKa = 4.33MCRR27 pKa = 11.84MVDD30 pKa = 3.21NLLSEE35 pKa = 4.43KK36 pKa = 10.39EE37 pKa = 4.13KK38 pKa = 10.52PDD40 pKa = 5.4PINQSYY46 pKa = 9.3PRR48 pKa = 11.84QSQEE52 pKa = 5.4DD53 pKa = 4.07EE54 pKa = 4.24NQPSDD59 pKa = 3.87TEE61 pKa = 4.44STEE64 pKa = 3.24SWTYY68 pKa = 11.24YY69 pKa = 10.74DD70 pKa = 4.12YY71 pKa = 11.21TDD73 pKa = 2.87HH74 pKa = 7.56RR75 pKa = 11.84YY76 pKa = 9.38YY77 pKa = 10.26PSYY80 pKa = 10.64EE81 pKa = 4.57DD82 pKa = 4.16NDD84 pKa = 3.78SDD86 pKa = 5.38IYY88 pKa = 11.69DD89 pKa = 4.05LMYY92 pKa = 10.64EE93 pKa = 4.44CNHH96 pKa = 6.31GEE98 pKa = 3.85WGNN101 pKa = 3.42
MM1 pKa = 7.18SASRR5 pKa = 11.84NTTTAPDD12 pKa = 3.52KK13 pKa = 10.79PGNQGDD19 pKa = 3.91EE20 pKa = 4.45IFIEE24 pKa = 4.33MCRR27 pKa = 11.84MVDD30 pKa = 3.21NLLSEE35 pKa = 4.43KK36 pKa = 10.39EE37 pKa = 4.13KK38 pKa = 10.52PDD40 pKa = 5.4PINQSYY46 pKa = 9.3PRR48 pKa = 11.84QSQEE52 pKa = 5.4DD53 pKa = 4.07EE54 pKa = 4.24NQPSDD59 pKa = 3.87TEE61 pKa = 4.44STEE64 pKa = 3.24SWTYY68 pKa = 11.24YY69 pKa = 10.74DD70 pKa = 4.12YY71 pKa = 11.21TDD73 pKa = 2.87HH74 pKa = 7.56RR75 pKa = 11.84YY76 pKa = 9.38YY77 pKa = 10.26PSYY80 pKa = 10.64EE81 pKa = 4.57DD82 pKa = 4.16NDD84 pKa = 3.78SDD86 pKa = 5.38IYY88 pKa = 11.69DD89 pKa = 4.05LMYY92 pKa = 10.64EE93 pKa = 4.44CNHH96 pKa = 6.31GEE98 pKa = 3.85WGNN101 pKa = 3.42
Molecular weight: 11.9 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1D8FVQ4|A0A1D8FVQ4_9RHAB Large structural protein OS=Physostegia chlorotic mottle virus OX=1905830 GN=L PE=4 SV=1
MM1 pKa = 7.46INVIGKK7 pKa = 8.02TNASVQGIYY16 pKa = 10.28EE17 pKa = 3.86VDD19 pKa = 3.56RR20 pKa = 11.84EE21 pKa = 4.41KK22 pKa = 11.31VSLLSPVTGQFVSLNYY38 pKa = 9.65IIKK41 pKa = 9.61INMYY45 pKa = 8.84EE46 pKa = 4.48KK47 pKa = 10.56EE48 pKa = 4.09MQKK51 pKa = 10.34SFEE54 pKa = 4.17EE55 pKa = 5.12GEE57 pKa = 4.17LHH59 pKa = 5.88YY60 pKa = 10.83HH61 pKa = 7.5DD62 pKa = 4.83IFDD65 pKa = 4.08MVEE68 pKa = 3.92SALNLPTTRR77 pKa = 11.84PDD79 pKa = 3.62SIKK82 pKa = 9.96RR83 pKa = 11.84TPTNLPTRR91 pKa = 11.84VAKK94 pKa = 9.57TIIQICRR101 pKa = 11.84IHH103 pKa = 7.41AIHH106 pKa = 6.74TNAKK110 pKa = 8.02MVFMTTSTDD119 pKa = 3.19HH120 pKa = 8.05SEE122 pKa = 4.11DD123 pKa = 3.74TPTLVIKK130 pKa = 10.57EE131 pKa = 4.35GEE133 pKa = 4.47TLVSPHH139 pKa = 6.58YY140 pKa = 9.59DD141 pKa = 3.03SHH143 pKa = 7.48VVEE146 pKa = 5.1VVNQPGEE153 pKa = 4.4SPLSGSYY160 pKa = 10.53HH161 pKa = 5.66LTLEE165 pKa = 4.41KK166 pKa = 10.41PYY168 pKa = 10.27ISQDD172 pKa = 3.01NKK174 pKa = 10.8IIGHH178 pKa = 6.99LSFAAYY184 pKa = 9.66VRR186 pKa = 11.84APPRR190 pKa = 11.84GNAVSQARR198 pKa = 11.84LNIRR202 pKa = 11.84VLASKK207 pKa = 8.1YY208 pKa = 8.7QRR210 pKa = 11.84SAAHH214 pKa = 6.57KK215 pKa = 10.27VNTLFRR221 pKa = 11.84KK222 pKa = 8.92SMKK225 pKa = 9.84RR226 pKa = 11.84KK227 pKa = 10.17SEE229 pKa = 4.05TSPTKK234 pKa = 10.63APSGSIMDD242 pKa = 4.37AVAKK246 pKa = 10.14ILKK249 pKa = 9.97QSS251 pKa = 3.17
MM1 pKa = 7.46INVIGKK7 pKa = 8.02TNASVQGIYY16 pKa = 10.28EE17 pKa = 3.86VDD19 pKa = 3.56RR20 pKa = 11.84EE21 pKa = 4.41KK22 pKa = 11.31VSLLSPVTGQFVSLNYY38 pKa = 9.65IIKK41 pKa = 9.61INMYY45 pKa = 8.84EE46 pKa = 4.48KK47 pKa = 10.56EE48 pKa = 4.09MQKK51 pKa = 10.34SFEE54 pKa = 4.17EE55 pKa = 5.12GEE57 pKa = 4.17LHH59 pKa = 5.88YY60 pKa = 10.83HH61 pKa = 7.5DD62 pKa = 4.83IFDD65 pKa = 4.08MVEE68 pKa = 3.92SALNLPTTRR77 pKa = 11.84PDD79 pKa = 3.62SIKK82 pKa = 9.96RR83 pKa = 11.84TPTNLPTRR91 pKa = 11.84VAKK94 pKa = 9.57TIIQICRR101 pKa = 11.84IHH103 pKa = 7.41AIHH106 pKa = 6.74TNAKK110 pKa = 8.02MVFMTTSTDD119 pKa = 3.19HH120 pKa = 8.05SEE122 pKa = 4.11DD123 pKa = 3.74TPTLVIKK130 pKa = 10.57EE131 pKa = 4.35GEE133 pKa = 4.47TLVSPHH139 pKa = 6.58YY140 pKa = 9.59DD141 pKa = 3.03SHH143 pKa = 7.48VVEE146 pKa = 5.1VVNQPGEE153 pKa = 4.4SPLSGSYY160 pKa = 10.53HH161 pKa = 5.66LTLEE165 pKa = 4.41KK166 pKa = 10.41PYY168 pKa = 10.27ISQDD172 pKa = 3.01NKK174 pKa = 10.8IIGHH178 pKa = 6.99LSFAAYY184 pKa = 9.66VRR186 pKa = 11.84APPRR190 pKa = 11.84GNAVSQARR198 pKa = 11.84LNIRR202 pKa = 11.84VLASKK207 pKa = 8.1YY208 pKa = 8.7QRR210 pKa = 11.84SAAHH214 pKa = 6.57KK215 pKa = 10.27VNTLFRR221 pKa = 11.84KK222 pKa = 8.92SMKK225 pKa = 9.84RR226 pKa = 11.84KK227 pKa = 10.17SEE229 pKa = 4.05TSPTKK234 pKa = 10.63APSGSIMDD242 pKa = 4.37AVAKK246 pKa = 10.14ILKK249 pKa = 9.97QSS251 pKa = 3.17
Molecular weight: 28.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3979 |
101 |
1946 |
568.4 |
64.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.976 ± 1.137 | 1.608 ± 0.398 |
6.132 ± 0.464 | 5.454 ± 0.535 |
3.418 ± 0.303 | 5.504 ± 0.352 |
2.312 ± 0.515 | 7.439 ± 0.384 |
5.604 ± 0.553 | 8.444 ± 0.994 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.971 ± 0.359 | 4.85 ± 0.649 |
5.077 ± 0.616 | 3.192 ± 0.279 |
4.775 ± 0.68 | 9.324 ± 0.626 |
6.032 ± 0.56 | 6.685 ± 0.505 |
1.206 ± 0.311 | 3.996 ± 0.607 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
