
Methylovulum psychrotolerans
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Methylococcales; Methylococcaceae; Methylovulum
Average proteome isoelectric point is 6.33
Get precalculated fractions of proteins
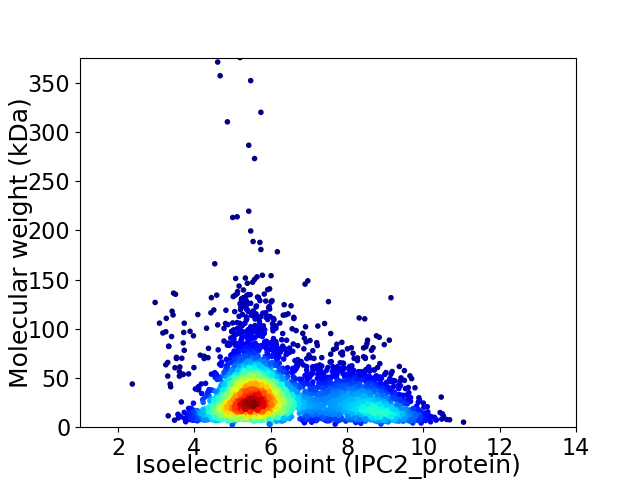
Virtual 2D-PAGE plot for 4299 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Z4C3I1|A0A1Z4C3I1_9GAMM Tyrosinase OS=Methylovulum psychrotolerans OX=1704499 GN=CEK71_19595 PE=4 SV=1
MM1 pKa = 7.54SNTAPSFITKK11 pKa = 9.86NDD13 pKa = 4.75PITTDD18 pKa = 4.2FGYY21 pKa = 10.62SDD23 pKa = 3.33QAYY26 pKa = 9.39DD27 pKa = 4.3VSVQTDD33 pKa = 3.51GKK35 pKa = 10.49LVISGTVYY43 pKa = 11.11LDD45 pKa = 3.0MMADD49 pKa = 3.58MGQGLMGSSVLRR61 pKa = 11.84YY62 pKa = 10.12NSDD65 pKa = 2.9GSLDD69 pKa = 3.74LSFSGDD75 pKa = 3.09GKK77 pKa = 11.0LDD79 pKa = 3.47TRR81 pKa = 11.84GLIHH85 pKa = 6.94AQALQADD92 pKa = 4.6GKK94 pKa = 10.65ILVAGTASTSFDD106 pKa = 3.88FYY108 pKa = 11.52DD109 pKa = 4.6FDD111 pKa = 3.91IEE113 pKa = 4.71RR114 pKa = 11.84YY115 pKa = 9.99NSDD118 pKa = 3.0GSLDD122 pKa = 3.6TTFGNGGVVTTNFLLSNDD140 pKa = 3.21IAYY143 pKa = 10.49SIAVQSDD150 pKa = 3.21GKK152 pKa = 10.49ILAAGKK158 pKa = 9.97SGHH161 pKa = 7.29DD162 pKa = 3.76YY163 pKa = 11.59GLVRR167 pKa = 11.84YY168 pKa = 9.02NSNGSLDD175 pKa = 3.62TSFSGDD181 pKa = 3.05GKK183 pKa = 10.86VATDD187 pKa = 3.41MGADD191 pKa = 4.17FEE193 pKa = 4.53QANSIVLQSNGKK205 pKa = 8.9IVLGGSSGLVRR216 pKa = 11.84YY217 pKa = 7.73NTDD220 pKa = 2.57GSLDD224 pKa = 3.51TGFSGDD230 pKa = 3.9GKK232 pKa = 10.7LSTSGITNLALQTDD246 pKa = 4.73GKK248 pKa = 10.94LLATEE253 pKa = 4.56ADD255 pKa = 3.85GTLDD259 pKa = 3.98RR260 pKa = 11.84YY261 pKa = 11.08NSDD264 pKa = 3.01GSVDD268 pKa = 3.46TSFSGDD274 pKa = 3.23GKK276 pKa = 9.95VTDD279 pKa = 3.39VWANTLLLQSDD290 pKa = 4.42GKK292 pKa = 11.32LLTTNNYY299 pKa = 10.13SEE301 pKa = 4.24LTRR304 pKa = 11.84YY305 pKa = 10.21NSDD308 pKa = 2.93GSLDD312 pKa = 3.59TSFSGDD318 pKa = 3.1GKK320 pKa = 9.12ITIGFTINATTVQADD335 pKa = 3.56GKK337 pKa = 10.54IVVTGYY343 pKa = 10.87GFNYY347 pKa = 8.0PQAYY351 pKa = 9.7LSYY354 pKa = 10.17YY355 pKa = 10.41RR356 pKa = 11.84YY357 pKa = 10.63DD358 pKa = 3.87FYY360 pKa = 10.9TARR363 pKa = 11.84YY364 pKa = 8.91NSDD367 pKa = 2.84GSLDD371 pKa = 3.65TTFTQSNDD379 pKa = 3.24SLRR382 pKa = 11.84LPLSITEE389 pKa = 4.45NDD391 pKa = 3.47PATYY395 pKa = 9.99LAEE398 pKa = 4.2LAQIRR403 pKa = 11.84DD404 pKa = 3.86TEE406 pKa = 4.2LDD408 pKa = 3.46ALNGGLGDD416 pKa = 3.68YY417 pKa = 10.75SGASLTLARR426 pKa = 11.84HH427 pKa = 6.09GGANTDD433 pKa = 3.8DD434 pKa = 3.76VFGGSGVLALSNGAITVSGVNIGTYY459 pKa = 7.39QQSGGQLTLTFGAATTQQVNSALQHH484 pKa = 5.21LTYY487 pKa = 10.83QNTSDD492 pKa = 4.05LTPPSLQIDD501 pKa = 3.54WTFSDD506 pKa = 4.46GALSTVGNTTVDD518 pKa = 3.17ISPQYY523 pKa = 11.04DD524 pKa = 3.25ATTGYY529 pKa = 11.88AMINGSRR536 pKa = 11.84TVGQTLTTSNNFTDD550 pKa = 4.07PDD552 pKa = 3.93GLGTISYY559 pKa = 9.0QWQGSSNGTTWSNLGSGPTLVVTQAMLSQQIRR591 pKa = 11.84LTDD594 pKa = 3.3SYY596 pKa = 11.45TDD598 pKa = 3.25QQGAAKK604 pKa = 10.41SITTVQGTSGNDD616 pKa = 3.24NILSGDD622 pKa = 3.71AKK624 pKa = 10.79FGMDD628 pKa = 4.72GNDD631 pKa = 3.14HH632 pKa = 6.93LYY634 pKa = 10.54SYY636 pKa = 11.04SSTYY640 pKa = 10.05GHH642 pKa = 6.53SNSILMDD649 pKa = 4.07GGNGKK654 pKa = 9.8DD655 pKa = 3.46VVDD658 pKa = 4.42YY659 pKa = 11.39SPADD663 pKa = 3.34ILNVGVTVDD672 pKa = 3.92LTLTTAQNTVAVGMNTLLNIEE693 pKa = 4.65DD694 pKa = 5.67LIGTLNNDD702 pKa = 3.44TFTGNALANSLFGQAGNDD720 pKa = 3.66TLNGGGGNDD729 pKa = 4.05GLTGGAGDD737 pKa = 4.61DD738 pKa = 4.04RR739 pKa = 11.84LDD741 pKa = 3.92GGTGTDD747 pKa = 2.94MAYY750 pKa = 10.72YY751 pKa = 10.19NSAATGVTVNLALTTAQNTHH771 pKa = 5.72GAGTDD776 pKa = 3.18TLLNIEE782 pKa = 4.54SLNGSTYY789 pKa = 11.14NDD791 pKa = 3.3VLVGNYY797 pKa = 10.24ANNTLLGGNGNDD809 pKa = 3.4VLAGSLGNDD818 pKa = 3.54TLTGGAGKK826 pKa = 10.2DD827 pKa = 2.69IFAFNSALGINNIDD841 pKa = 3.59TLTDD845 pKa = 3.58FNVADD850 pKa = 3.6DD851 pKa = 4.9TIRR854 pKa = 11.84LEE856 pKa = 4.07NGIFTTLTTPGVLAAEE872 pKa = 4.21AFKK875 pKa = 10.65IIGNGGVADD884 pKa = 4.05STDD887 pKa = 3.0HH888 pKa = 6.96LLYY891 pKa = 9.91NTATGALSYY900 pKa = 11.0DD901 pKa = 3.68ADD903 pKa = 3.99GSGAAAAVQIAILGTGLAMTNADD926 pKa = 4.33FIVVV930 pKa = 3.78
MM1 pKa = 7.54SNTAPSFITKK11 pKa = 9.86NDD13 pKa = 4.75PITTDD18 pKa = 4.2FGYY21 pKa = 10.62SDD23 pKa = 3.33QAYY26 pKa = 9.39DD27 pKa = 4.3VSVQTDD33 pKa = 3.51GKK35 pKa = 10.49LVISGTVYY43 pKa = 11.11LDD45 pKa = 3.0MMADD49 pKa = 3.58MGQGLMGSSVLRR61 pKa = 11.84YY62 pKa = 10.12NSDD65 pKa = 2.9GSLDD69 pKa = 3.74LSFSGDD75 pKa = 3.09GKK77 pKa = 11.0LDD79 pKa = 3.47TRR81 pKa = 11.84GLIHH85 pKa = 6.94AQALQADD92 pKa = 4.6GKK94 pKa = 10.65ILVAGTASTSFDD106 pKa = 3.88FYY108 pKa = 11.52DD109 pKa = 4.6FDD111 pKa = 3.91IEE113 pKa = 4.71RR114 pKa = 11.84YY115 pKa = 9.99NSDD118 pKa = 3.0GSLDD122 pKa = 3.6TTFGNGGVVTTNFLLSNDD140 pKa = 3.21IAYY143 pKa = 10.49SIAVQSDD150 pKa = 3.21GKK152 pKa = 10.49ILAAGKK158 pKa = 9.97SGHH161 pKa = 7.29DD162 pKa = 3.76YY163 pKa = 11.59GLVRR167 pKa = 11.84YY168 pKa = 9.02NSNGSLDD175 pKa = 3.62TSFSGDD181 pKa = 3.05GKK183 pKa = 10.86VATDD187 pKa = 3.41MGADD191 pKa = 4.17FEE193 pKa = 4.53QANSIVLQSNGKK205 pKa = 8.9IVLGGSSGLVRR216 pKa = 11.84YY217 pKa = 7.73NTDD220 pKa = 2.57GSLDD224 pKa = 3.51TGFSGDD230 pKa = 3.9GKK232 pKa = 10.7LSTSGITNLALQTDD246 pKa = 4.73GKK248 pKa = 10.94LLATEE253 pKa = 4.56ADD255 pKa = 3.85GTLDD259 pKa = 3.98RR260 pKa = 11.84YY261 pKa = 11.08NSDD264 pKa = 3.01GSVDD268 pKa = 3.46TSFSGDD274 pKa = 3.23GKK276 pKa = 9.95VTDD279 pKa = 3.39VWANTLLLQSDD290 pKa = 4.42GKK292 pKa = 11.32LLTTNNYY299 pKa = 10.13SEE301 pKa = 4.24LTRR304 pKa = 11.84YY305 pKa = 10.21NSDD308 pKa = 2.93GSLDD312 pKa = 3.59TSFSGDD318 pKa = 3.1GKK320 pKa = 9.12ITIGFTINATTVQADD335 pKa = 3.56GKK337 pKa = 10.54IVVTGYY343 pKa = 10.87GFNYY347 pKa = 8.0PQAYY351 pKa = 9.7LSYY354 pKa = 10.17YY355 pKa = 10.41RR356 pKa = 11.84YY357 pKa = 10.63DD358 pKa = 3.87FYY360 pKa = 10.9TARR363 pKa = 11.84YY364 pKa = 8.91NSDD367 pKa = 2.84GSLDD371 pKa = 3.65TTFTQSNDD379 pKa = 3.24SLRR382 pKa = 11.84LPLSITEE389 pKa = 4.45NDD391 pKa = 3.47PATYY395 pKa = 9.99LAEE398 pKa = 4.2LAQIRR403 pKa = 11.84DD404 pKa = 3.86TEE406 pKa = 4.2LDD408 pKa = 3.46ALNGGLGDD416 pKa = 3.68YY417 pKa = 10.75SGASLTLARR426 pKa = 11.84HH427 pKa = 6.09GGANTDD433 pKa = 3.8DD434 pKa = 3.76VFGGSGVLALSNGAITVSGVNIGTYY459 pKa = 7.39QQSGGQLTLTFGAATTQQVNSALQHH484 pKa = 5.21LTYY487 pKa = 10.83QNTSDD492 pKa = 4.05LTPPSLQIDD501 pKa = 3.54WTFSDD506 pKa = 4.46GALSTVGNTTVDD518 pKa = 3.17ISPQYY523 pKa = 11.04DD524 pKa = 3.25ATTGYY529 pKa = 11.88AMINGSRR536 pKa = 11.84TVGQTLTTSNNFTDD550 pKa = 4.07PDD552 pKa = 3.93GLGTISYY559 pKa = 9.0QWQGSSNGTTWSNLGSGPTLVVTQAMLSQQIRR591 pKa = 11.84LTDD594 pKa = 3.3SYY596 pKa = 11.45TDD598 pKa = 3.25QQGAAKK604 pKa = 10.41SITTVQGTSGNDD616 pKa = 3.24NILSGDD622 pKa = 3.71AKK624 pKa = 10.79FGMDD628 pKa = 4.72GNDD631 pKa = 3.14HH632 pKa = 6.93LYY634 pKa = 10.54SYY636 pKa = 11.04SSTYY640 pKa = 10.05GHH642 pKa = 6.53SNSILMDD649 pKa = 4.07GGNGKK654 pKa = 9.8DD655 pKa = 3.46VVDD658 pKa = 4.42YY659 pKa = 11.39SPADD663 pKa = 3.34ILNVGVTVDD672 pKa = 3.92LTLTTAQNTVAVGMNTLLNIEE693 pKa = 4.65DD694 pKa = 5.67LIGTLNNDD702 pKa = 3.44TFTGNALANSLFGQAGNDD720 pKa = 3.66TLNGGGGNDD729 pKa = 4.05GLTGGAGDD737 pKa = 4.61DD738 pKa = 4.04RR739 pKa = 11.84LDD741 pKa = 3.92GGTGTDD747 pKa = 2.94MAYY750 pKa = 10.72YY751 pKa = 10.19NSAATGVTVNLALTTAQNTHH771 pKa = 5.72GAGTDD776 pKa = 3.18TLLNIEE782 pKa = 4.54SLNGSTYY789 pKa = 11.14NDD791 pKa = 3.3VLVGNYY797 pKa = 10.24ANNTLLGGNGNDD809 pKa = 3.4VLAGSLGNDD818 pKa = 3.54TLTGGAGKK826 pKa = 10.2DD827 pKa = 2.69IFAFNSALGINNIDD841 pKa = 3.59TLTDD845 pKa = 3.58FNVADD850 pKa = 3.6DD851 pKa = 4.9TIRR854 pKa = 11.84LEE856 pKa = 4.07NGIFTTLTTPGVLAAEE872 pKa = 4.21AFKK875 pKa = 10.65IIGNGGVADD884 pKa = 4.05STDD887 pKa = 3.0HH888 pKa = 6.96LLYY891 pKa = 9.91NTATGALSYY900 pKa = 11.0DD901 pKa = 3.68ADD903 pKa = 3.99GSGAAAAVQIAILGTGLAMTNADD926 pKa = 4.33FIVVV930 pKa = 3.78
Molecular weight: 96.31 kDa
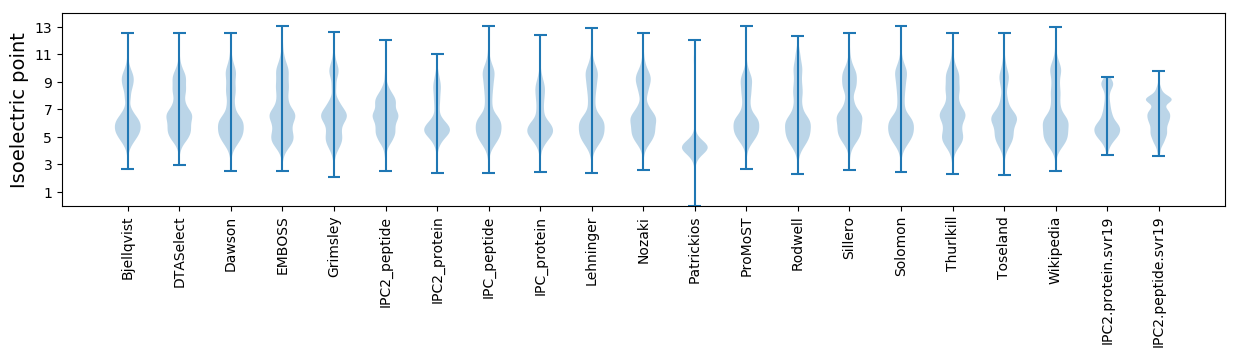
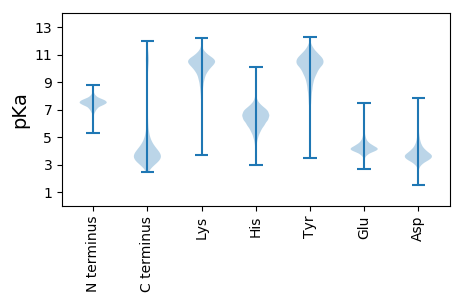
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Z4BVX5|A0A1Z4BVX5_9GAMM Uncharacterized protein OS=Methylovulum psychrotolerans OX=1704499 GN=CEK71_04840 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.17QPSKK9 pKa = 8.35IKK11 pKa = 10.26RR12 pKa = 11.84VRR14 pKa = 11.84THH16 pKa = 5.82GFRR19 pKa = 11.84SRR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.1VLNSRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.18GRR39 pKa = 11.84HH40 pKa = 5.12KK41 pKa = 10.72LALL44 pKa = 3.92
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.17QPSKK9 pKa = 8.35IKK11 pKa = 10.26RR12 pKa = 11.84VRR14 pKa = 11.84THH16 pKa = 5.82GFRR19 pKa = 11.84SRR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.1VLNSRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.18GRR39 pKa = 11.84HH40 pKa = 5.12KK41 pKa = 10.72LALL44 pKa = 3.92
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1384159 |
18 |
3606 |
322.0 |
35.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.419 ± 0.049 | 1.07 ± 0.013 |
5.539 ± 0.032 | 5.263 ± 0.048 |
3.94 ± 0.024 | 7.265 ± 0.054 |
2.364 ± 0.021 | 5.76 ± 0.032 |
4.634 ± 0.04 | 11.174 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.145 ± 0.017 | 3.945 ± 0.032 |
4.578 ± 0.032 | 4.741 ± 0.034 |
4.883 ± 0.035 | 5.73 ± 0.034 |
5.554 ± 0.038 | 6.57 ± 0.03 |
1.33 ± 0.018 | 3.095 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
