
Kwoniella pini CBS 10737
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Tremellomycetes; Tremellales; Cryptococcaceae; Kwoniella; Kwoniella pini
Average proteome isoelectric point is 6.5
Get precalculated fractions of proteins
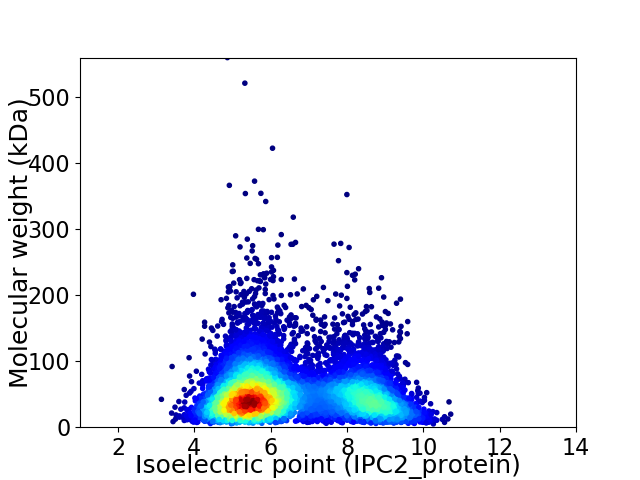
Virtual 2D-PAGE plot for 7827 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B9ICF3|A0A1B9ICF3_9TREE Prephenate dehydrogenase [NADP(+)] OS=Kwoniella pini CBS 10737 OX=1296096 GN=I206_00415 PE=3 SV=1
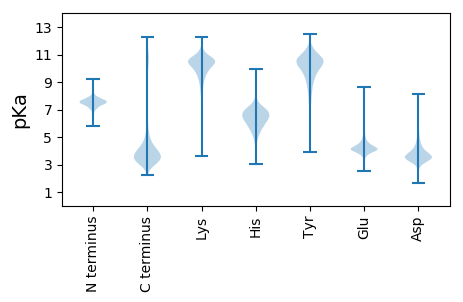
MM1 pKa = 7.27FSKK4 pKa = 10.84ALFLLPLLPLLSAAPLPYY22 pKa = 9.47PLLGAGPISDD32 pKa = 4.74LDD34 pKa = 3.94SVINPLTQDD43 pKa = 3.75DD44 pKa = 4.88NLPGLPVTPGDD55 pKa = 3.96PSPTLSNTGDD65 pKa = 3.27GSLINGLLDD74 pKa = 3.49TVEE77 pKa = 4.76TITGPLNVSIGANVDD92 pKa = 3.68LLGINTTVGLNVDD105 pKa = 4.14LDD107 pKa = 4.19DD108 pKa = 5.52DD109 pKa = 4.35EE110 pKa = 6.09EE111 pKa = 4.62MICGPVNGYY120 pKa = 8.61WSNQQYY126 pKa = 10.87NIPCACWSDD135 pKa = 3.38SRR137 pKa = 11.84GLVISAQLEE146 pKa = 4.01ADD148 pKa = 3.67LGLDD152 pKa = 3.35QVEE155 pKa = 4.41GLDD158 pKa = 3.79TFLQAQIEE166 pKa = 4.45FGGEE170 pKa = 3.62RR171 pKa = 11.84FTYY174 pKa = 9.36PAYY177 pKa = 11.21SMPTCDD183 pKa = 4.14GDD185 pKa = 4.7GGFNCPGGRR194 pKa = 11.84SSNGKK199 pKa = 9.43CSKK202 pKa = 10.37FLAAKK207 pKa = 8.78PRR209 pKa = 11.84PKK211 pKa = 10.72VLIQSTSASPASVPTANSVPTATDD235 pKa = 3.17AVINEE240 pKa = 4.3VPPTTLNSIPTSTIAGFDD258 pKa = 3.44NVQPTVTPITTVSSPDD274 pKa = 2.99QTQPVAEE281 pKa = 4.21SQEE284 pKa = 3.91VDD286 pKa = 4.0DD287 pKa = 5.34EE288 pKa = 5.12SISADD293 pKa = 3.19NVVLTTSTSTSTVILPATVFVEE315 pKa = 4.55MVTTTQPTTIWATEE329 pKa = 4.23TQTQTQTQTQTQTQTITSTSVQTQWATQTQWATTTLSNCAANDD372 pKa = 3.26EE373 pKa = 4.67EE374 pKa = 5.2INVNSVSQSVQEE386 pKa = 4.24AGYY389 pKa = 8.35TPTPTSTYY397 pKa = 8.27TSSSISSTISSSSSISTSSSTVSQVLPTPSINNSVPTQTSNNTGDD442 pKa = 3.98EE443 pKa = 4.17DD444 pKa = 5.92DD445 pKa = 5.39EE446 pKa = 6.05EE447 pKa = 6.71DD448 pKa = 4.57DD449 pKa = 3.58QFKK452 pKa = 10.59PGGVIKK458 pKa = 10.73LSHH461 pKa = 7.21PPLKK465 pKa = 10.8SQLKK469 pKa = 9.25CSNGEE474 pKa = 3.91EE475 pKa = 4.75FKK477 pKa = 8.72TTMCCRR483 pKa = 11.84VDD485 pKa = 3.71QIEE488 pKa = 4.41LNGEE492 pKa = 4.43CKK494 pKa = 10.0CSKK497 pKa = 10.54GFEE500 pKa = 4.15NVLNLNLCLSICLGNRR516 pKa = 11.84LPSGEE521 pKa = 4.72CSLLDD526 pKa = 4.52LNTNLDD532 pKa = 4.0LGLSDD537 pKa = 4.55ILSPLL542 pKa = 3.49
MM1 pKa = 7.27FSKK4 pKa = 10.84ALFLLPLLPLLSAAPLPYY22 pKa = 9.47PLLGAGPISDD32 pKa = 4.74LDD34 pKa = 3.94SVINPLTQDD43 pKa = 3.75DD44 pKa = 4.88NLPGLPVTPGDD55 pKa = 3.96PSPTLSNTGDD65 pKa = 3.27GSLINGLLDD74 pKa = 3.49TVEE77 pKa = 4.76TITGPLNVSIGANVDD92 pKa = 3.68LLGINTTVGLNVDD105 pKa = 4.14LDD107 pKa = 4.19DD108 pKa = 5.52DD109 pKa = 4.35EE110 pKa = 6.09EE111 pKa = 4.62MICGPVNGYY120 pKa = 8.61WSNQQYY126 pKa = 10.87NIPCACWSDD135 pKa = 3.38SRR137 pKa = 11.84GLVISAQLEE146 pKa = 4.01ADD148 pKa = 3.67LGLDD152 pKa = 3.35QVEE155 pKa = 4.41GLDD158 pKa = 3.79TFLQAQIEE166 pKa = 4.45FGGEE170 pKa = 3.62RR171 pKa = 11.84FTYY174 pKa = 9.36PAYY177 pKa = 11.21SMPTCDD183 pKa = 4.14GDD185 pKa = 4.7GGFNCPGGRR194 pKa = 11.84SSNGKK199 pKa = 9.43CSKK202 pKa = 10.37FLAAKK207 pKa = 8.78PRR209 pKa = 11.84PKK211 pKa = 10.72VLIQSTSASPASVPTANSVPTATDD235 pKa = 3.17AVINEE240 pKa = 4.3VPPTTLNSIPTSTIAGFDD258 pKa = 3.44NVQPTVTPITTVSSPDD274 pKa = 2.99QTQPVAEE281 pKa = 4.21SQEE284 pKa = 3.91VDD286 pKa = 4.0DD287 pKa = 5.34EE288 pKa = 5.12SISADD293 pKa = 3.19NVVLTTSTSTSTVILPATVFVEE315 pKa = 4.55MVTTTQPTTIWATEE329 pKa = 4.23TQTQTQTQTQTQTQTITSTSVQTQWATQTQWATTTLSNCAANDD372 pKa = 3.26EE373 pKa = 4.67EE374 pKa = 5.2INVNSVSQSVQEE386 pKa = 4.24AGYY389 pKa = 8.35TPTPTSTYY397 pKa = 8.27TSSSISSTISSSSSISTSSSTVSQVLPTPSINNSVPTQTSNNTGDD442 pKa = 3.98EE443 pKa = 4.17DD444 pKa = 5.92DD445 pKa = 5.39EE446 pKa = 6.05EE447 pKa = 6.71DD448 pKa = 4.57DD449 pKa = 3.58QFKK452 pKa = 10.59PGGVIKK458 pKa = 10.73LSHH461 pKa = 7.21PPLKK465 pKa = 10.8SQLKK469 pKa = 9.25CSNGEE474 pKa = 3.91EE475 pKa = 4.75FKK477 pKa = 8.72TTMCCRR483 pKa = 11.84VDD485 pKa = 3.71QIEE488 pKa = 4.41LNGEE492 pKa = 4.43CKK494 pKa = 10.0CSKK497 pKa = 10.54GFEE500 pKa = 4.15NVLNLNLCLSICLGNRR516 pKa = 11.84LPSGEE521 pKa = 4.72CSLLDD526 pKa = 4.52LNTNLDD532 pKa = 4.0LGLSDD537 pKa = 4.55ILSPLL542 pKa = 3.49
Molecular weight: 57.04 kDa
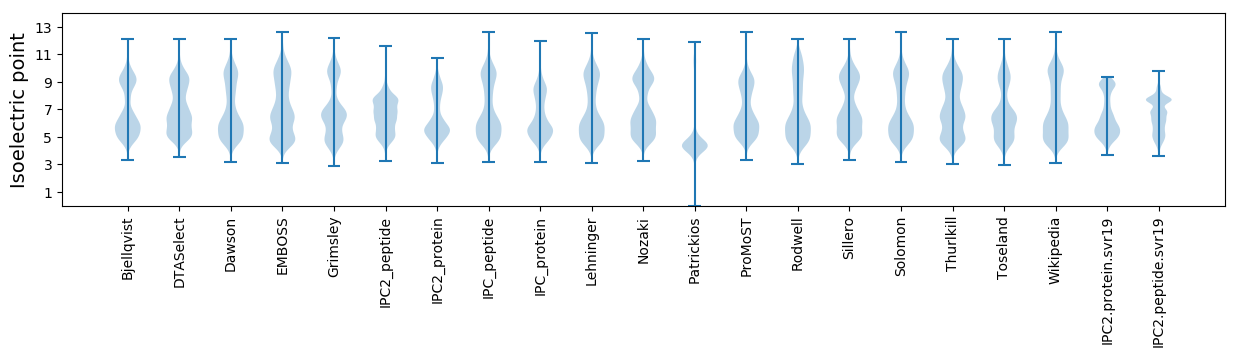
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B9I632|A0A1B9I632_9TREE YbgI/family dinuclear metal center protein OS=Kwoniella pini CBS 10737 OX=1296096 GN=I206_03045 PE=3 SV=1
MM1 pKa = 7.82PSLHH5 pKa = 6.17QLCPDD10 pKa = 3.97CGLPRR15 pKa = 11.84ASPPPPLDD23 pKa = 3.27TMFEE27 pKa = 4.18EE28 pKa = 4.77DD29 pKa = 4.18FEE31 pKa = 5.16RR32 pKa = 11.84CFICEE37 pKa = 4.09KK38 pKa = 10.24ACKK41 pKa = 9.98GLYY44 pKa = 9.87CSSEE48 pKa = 3.78CRR50 pKa = 11.84LRR52 pKa = 11.84DD53 pKa = 3.24QGTPSPAIRR62 pKa = 11.84ANRR65 pKa = 11.84GPVKK69 pKa = 9.73ITSQLPAALSPQVRR83 pKa = 11.84ATQNIGRR90 pKa = 11.84SPRR93 pKa = 11.84VQPQNRR99 pKa = 11.84GSSSISSGSSVSSSPLQSPQTNPSEE124 pKa = 4.16ADD126 pKa = 3.22SPKK129 pKa = 10.18RR130 pKa = 11.84DD131 pKa = 3.86NFDD134 pKa = 4.39LPPPAYY140 pKa = 7.12PTKK143 pKa = 10.75CFGILPASVPMKK155 pKa = 10.46IPALTARR162 pKa = 11.84ASPLVAPSQTPGSHH176 pKa = 6.53GSTVYY181 pKa = 10.26PAGASIDD188 pKa = 3.52TLRR191 pKa = 11.84FGRR194 pKa = 11.84KK195 pKa = 7.94PSAVNTVLSPNALIPRR211 pKa = 11.84CACGKK216 pKa = 8.99PANHH220 pKa = 7.31RR221 pKa = 11.84NRR223 pKa = 11.84GSSKK227 pKa = 10.71DD228 pKa = 3.5RR229 pKa = 11.84AEE231 pKa = 4.23LADD234 pKa = 3.68SGFSRR239 pKa = 11.84LSLGPSVISAPHH251 pKa = 5.78AQEE254 pKa = 4.16EE255 pKa = 4.43PNPRR259 pKa = 11.84SVRR262 pKa = 11.84IVSEE266 pKa = 4.0SSLPGRR272 pKa = 11.84PYY274 pKa = 10.88QNGLGTPGRR283 pKa = 11.84TALPLGIPSSPQVTASTSFLSRR305 pKa = 11.84SRR307 pKa = 11.84SDD309 pKa = 4.62PIPSSLMAQRR319 pKa = 11.84KK320 pKa = 8.89AIPAAPAPAPLITNVITPSHH340 pKa = 6.85RR341 pKa = 11.84EE342 pKa = 3.75LSEE345 pKa = 4.21MPLSPIVPALGRR357 pKa = 11.84PSRR360 pKa = 11.84SKK362 pKa = 10.22IALDD366 pKa = 3.32VDD368 pKa = 4.2VNSPRR373 pKa = 11.84RR374 pKa = 11.84GRR376 pKa = 11.84SRR378 pKa = 11.84EE379 pKa = 3.92RR380 pKa = 11.84QEE382 pKa = 3.92HH383 pKa = 6.26HH384 pKa = 6.37VGQMTSNFGGPADD397 pKa = 4.34RR398 pKa = 11.84EE399 pKa = 4.26QAPSRR404 pKa = 11.84SRR406 pKa = 11.84TRR408 pKa = 11.84RR409 pKa = 11.84EE410 pKa = 3.29SRR412 pKa = 11.84RR413 pKa = 11.84RR414 pKa = 11.84SDD416 pKa = 2.83SRR418 pKa = 11.84NKK420 pKa = 9.0EE421 pKa = 3.64RR422 pKa = 11.84SRR424 pKa = 11.84GGSGRR429 pKa = 11.84PSRR432 pKa = 11.84EE433 pKa = 3.39RR434 pKa = 11.84PIEE437 pKa = 4.27EE438 pKa = 4.2IEE440 pKa = 4.12RR441 pKa = 11.84TGQRR445 pKa = 11.84SPIQQQLNSPQILPSWSRR463 pKa = 11.84RR464 pKa = 11.84ASEE467 pKa = 3.73ATADD471 pKa = 3.72RR472 pKa = 11.84RR473 pKa = 11.84RR474 pKa = 11.84VLGEE478 pKa = 3.5AAPAMRR484 pKa = 11.84RR485 pKa = 11.84TASGGKK491 pKa = 9.5KK492 pKa = 10.23SPICEE497 pKa = 3.89RR498 pKa = 11.84GRR500 pKa = 11.84EE501 pKa = 3.85RR502 pKa = 11.84DD503 pKa = 3.28KK504 pKa = 11.55DD505 pKa = 3.37PEE507 pKa = 4.03EE508 pKa = 4.11RR509 pKa = 11.84KK510 pKa = 9.85KK511 pKa = 11.3EE512 pKa = 4.01EE513 pKa = 4.18LDD515 pKa = 3.47RR516 pKa = 11.84TSKK519 pKa = 10.6QLSQVFGVAAVV530 pKa = 3.23
MM1 pKa = 7.82PSLHH5 pKa = 6.17QLCPDD10 pKa = 3.97CGLPRR15 pKa = 11.84ASPPPPLDD23 pKa = 3.27TMFEE27 pKa = 4.18EE28 pKa = 4.77DD29 pKa = 4.18FEE31 pKa = 5.16RR32 pKa = 11.84CFICEE37 pKa = 4.09KK38 pKa = 10.24ACKK41 pKa = 9.98GLYY44 pKa = 9.87CSSEE48 pKa = 3.78CRR50 pKa = 11.84LRR52 pKa = 11.84DD53 pKa = 3.24QGTPSPAIRR62 pKa = 11.84ANRR65 pKa = 11.84GPVKK69 pKa = 9.73ITSQLPAALSPQVRR83 pKa = 11.84ATQNIGRR90 pKa = 11.84SPRR93 pKa = 11.84VQPQNRR99 pKa = 11.84GSSSISSGSSVSSSPLQSPQTNPSEE124 pKa = 4.16ADD126 pKa = 3.22SPKK129 pKa = 10.18RR130 pKa = 11.84DD131 pKa = 3.86NFDD134 pKa = 4.39LPPPAYY140 pKa = 7.12PTKK143 pKa = 10.75CFGILPASVPMKK155 pKa = 10.46IPALTARR162 pKa = 11.84ASPLVAPSQTPGSHH176 pKa = 6.53GSTVYY181 pKa = 10.26PAGASIDD188 pKa = 3.52TLRR191 pKa = 11.84FGRR194 pKa = 11.84KK195 pKa = 7.94PSAVNTVLSPNALIPRR211 pKa = 11.84CACGKK216 pKa = 8.99PANHH220 pKa = 7.31RR221 pKa = 11.84NRR223 pKa = 11.84GSSKK227 pKa = 10.71DD228 pKa = 3.5RR229 pKa = 11.84AEE231 pKa = 4.23LADD234 pKa = 3.68SGFSRR239 pKa = 11.84LSLGPSVISAPHH251 pKa = 5.78AQEE254 pKa = 4.16EE255 pKa = 4.43PNPRR259 pKa = 11.84SVRR262 pKa = 11.84IVSEE266 pKa = 4.0SSLPGRR272 pKa = 11.84PYY274 pKa = 10.88QNGLGTPGRR283 pKa = 11.84TALPLGIPSSPQVTASTSFLSRR305 pKa = 11.84SRR307 pKa = 11.84SDD309 pKa = 4.62PIPSSLMAQRR319 pKa = 11.84KK320 pKa = 8.89AIPAAPAPAPLITNVITPSHH340 pKa = 6.85RR341 pKa = 11.84EE342 pKa = 3.75LSEE345 pKa = 4.21MPLSPIVPALGRR357 pKa = 11.84PSRR360 pKa = 11.84SKK362 pKa = 10.22IALDD366 pKa = 3.32VDD368 pKa = 4.2VNSPRR373 pKa = 11.84RR374 pKa = 11.84GRR376 pKa = 11.84SRR378 pKa = 11.84EE379 pKa = 3.92RR380 pKa = 11.84QEE382 pKa = 3.92HH383 pKa = 6.26HH384 pKa = 6.37VGQMTSNFGGPADD397 pKa = 4.34RR398 pKa = 11.84EE399 pKa = 4.26QAPSRR404 pKa = 11.84SRR406 pKa = 11.84TRR408 pKa = 11.84RR409 pKa = 11.84EE410 pKa = 3.29SRR412 pKa = 11.84RR413 pKa = 11.84RR414 pKa = 11.84SDD416 pKa = 2.83SRR418 pKa = 11.84NKK420 pKa = 9.0EE421 pKa = 3.64RR422 pKa = 11.84SRR424 pKa = 11.84GGSGRR429 pKa = 11.84PSRR432 pKa = 11.84EE433 pKa = 3.39RR434 pKa = 11.84PIEE437 pKa = 4.27EE438 pKa = 4.2IEE440 pKa = 4.12RR441 pKa = 11.84TGQRR445 pKa = 11.84SPIQQQLNSPQILPSWSRR463 pKa = 11.84RR464 pKa = 11.84ASEE467 pKa = 3.73ATADD471 pKa = 3.72RR472 pKa = 11.84RR473 pKa = 11.84RR474 pKa = 11.84VLGEE478 pKa = 3.5AAPAMRR484 pKa = 11.84RR485 pKa = 11.84TASGGKK491 pKa = 9.5KK492 pKa = 10.23SPICEE497 pKa = 3.89RR498 pKa = 11.84GRR500 pKa = 11.84EE501 pKa = 3.85RR502 pKa = 11.84DD503 pKa = 3.28KK504 pKa = 11.55DD505 pKa = 3.37PEE507 pKa = 4.03EE508 pKa = 4.11RR509 pKa = 11.84KK510 pKa = 9.85KK511 pKa = 11.3EE512 pKa = 4.01EE513 pKa = 4.18LDD515 pKa = 3.47RR516 pKa = 11.84TSKK519 pKa = 10.6QLSQVFGVAAVV530 pKa = 3.23
Molecular weight: 57.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4033902 |
49 |
5028 |
515.4 |
57.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.009 ± 0.027 | 0.957 ± 0.01 |
5.488 ± 0.019 | 6.519 ± 0.029 |
3.456 ± 0.016 | 6.708 ± 0.027 |
2.195 ± 0.011 | 5.915 ± 0.023 |
5.788 ± 0.029 | 8.868 ± 0.03 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.964 ± 0.009 | 4.68 ± 0.021 |
6.232 ± 0.03 | 3.983 ± 0.02 |
5.378 ± 0.02 | 9.676 ± 0.041 |
5.961 ± 0.019 | 5.271 ± 0.021 |
1.323 ± 0.009 | 2.627 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
