
Capybara microvirus Cap3_SP_546
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.04
Get precalculated fractions of proteins
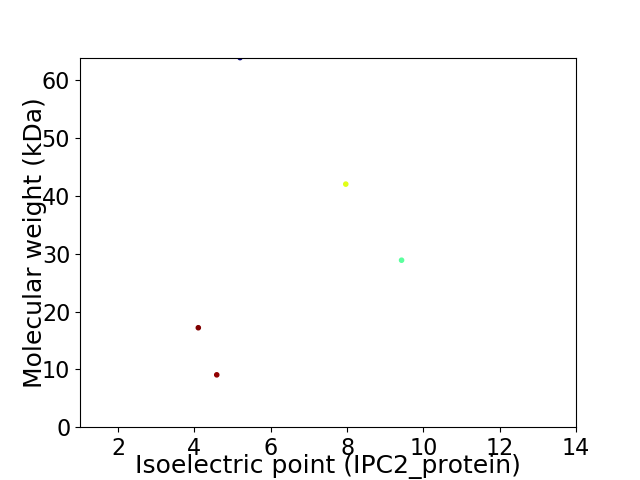
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W6C6|A0A4P8W6C6_9VIRU Replication initiator protein OS=Capybara microvirus Cap3_SP_546 OX=2585473 PE=4 SV=1
MM1 pKa = 7.42FRR3 pKa = 11.84TKK5 pKa = 10.63YY6 pKa = 10.49NCDD9 pKa = 3.15NSSVVSNNGSPFIDD23 pKa = 3.39TYY25 pKa = 11.78GFTTDD30 pKa = 4.91DD31 pKa = 3.65YY32 pKa = 11.0GNTIVGVTGQSNLFDD47 pKa = 5.52IIQSNSDD54 pKa = 3.33SGNISLIVDD63 pKa = 3.99RR64 pKa = 11.84ATRR67 pKa = 11.84GDD69 pKa = 3.99LSYY72 pKa = 11.67LNAVNSAYY80 pKa = 10.49FDD82 pKa = 4.03ARR84 pKa = 11.84DD85 pKa = 3.46LPTSYY90 pKa = 11.01RR91 pKa = 11.84EE92 pKa = 3.49MTEE95 pKa = 4.45YY96 pKa = 10.48INHH99 pKa = 6.7CNNTFNEE106 pKa = 4.29LPAHH110 pKa = 6.22IKK112 pKa = 9.95EE113 pKa = 4.33AFNNSSSEE121 pKa = 4.0FWSTFGSDD129 pKa = 2.25SWNRR133 pKa = 11.84RR134 pKa = 11.84VGFNSEE140 pKa = 4.44DD141 pKa = 3.49SSSNDD146 pKa = 2.81GGDD149 pKa = 3.63FNEE152 pKa = 4.56QEE154 pKa = 4.07
MM1 pKa = 7.42FRR3 pKa = 11.84TKK5 pKa = 10.63YY6 pKa = 10.49NCDD9 pKa = 3.15NSSVVSNNGSPFIDD23 pKa = 3.39TYY25 pKa = 11.78GFTTDD30 pKa = 4.91DD31 pKa = 3.65YY32 pKa = 11.0GNTIVGVTGQSNLFDD47 pKa = 5.52IIQSNSDD54 pKa = 3.33SGNISLIVDD63 pKa = 3.99RR64 pKa = 11.84ATRR67 pKa = 11.84GDD69 pKa = 3.99LSYY72 pKa = 11.67LNAVNSAYY80 pKa = 10.49FDD82 pKa = 4.03ARR84 pKa = 11.84DD85 pKa = 3.46LPTSYY90 pKa = 11.01RR91 pKa = 11.84EE92 pKa = 3.49MTEE95 pKa = 4.45YY96 pKa = 10.48INHH99 pKa = 6.7CNNTFNEE106 pKa = 4.29LPAHH110 pKa = 6.22IKK112 pKa = 9.95EE113 pKa = 4.33AFNNSSSEE121 pKa = 4.0FWSTFGSDD129 pKa = 2.25SWNRR133 pKa = 11.84RR134 pKa = 11.84VGFNSEE140 pKa = 4.44DD141 pKa = 3.49SSSNDD146 pKa = 2.81GGDD149 pKa = 3.63FNEE152 pKa = 4.56QEE154 pKa = 4.07
Molecular weight: 17.21 kDa
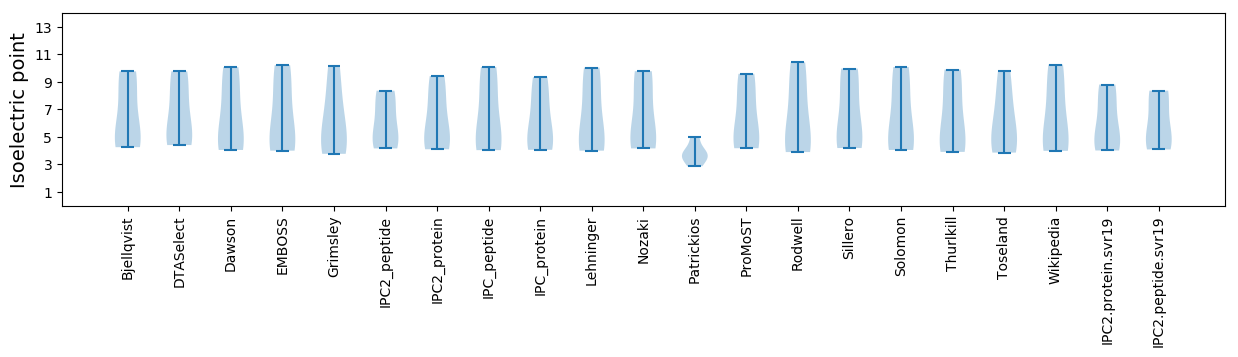
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W8U1|A0A4P8W8U1_9VIRU Major capsid protein OS=Capybara microvirus Cap3_SP_546 OX=2585473 PE=3 SV=1
MM1 pKa = 7.36SAYY4 pKa = 9.44TEE6 pKa = 4.22ALQLQRR12 pKa = 11.84DD13 pKa = 3.89AQSFTKK19 pKa = 9.66QQWNFEE25 pKa = 4.33QNLSSTAHH33 pKa = 4.86QRR35 pKa = 11.84EE36 pKa = 4.02VSDD39 pKa = 4.25LKK41 pKa = 11.07KK42 pKa = 10.88AGLNPVLSANSGASTPSVGLDD63 pKa = 3.11SGVQAASGYY72 pKa = 9.06MSSKK76 pKa = 8.64TAAAATRR83 pKa = 11.84YY84 pKa = 9.19ASNMSYY90 pKa = 10.81KK91 pKa = 10.49AALAQASATRR101 pKa = 11.84YY102 pKa = 8.86AASLNYY108 pKa = 8.26EE109 pKa = 4.13AQMSSNKK116 pKa = 10.16YY117 pKa = 9.37GIAMQIANSAAGEE130 pKa = 4.16GGVIGFIKK138 pKa = 10.34RR139 pKa = 11.84AVNSAKK145 pKa = 10.38SYY147 pKa = 11.0NSSIDD152 pKa = 3.41NTVKK156 pKa = 10.77SNNISIPNVKK166 pKa = 8.45VTPQSVLNGKK176 pKa = 8.9ALNGVKK182 pKa = 10.11RR183 pKa = 11.84EE184 pKa = 4.14DD185 pKa = 3.48LLKK188 pKa = 10.46PVTSSSQYY196 pKa = 9.7QFGNKK201 pKa = 6.86VTLNDD206 pKa = 3.32TGKK209 pKa = 10.59KK210 pKa = 9.33YY211 pKa = 10.63VSKK214 pKa = 10.73VLSDD218 pKa = 4.01LGVSTKK224 pKa = 10.87APIGKK229 pKa = 8.78EE230 pKa = 3.82CQIYY234 pKa = 8.88LQQYY238 pKa = 9.52LITGNLVYY246 pKa = 10.82YY247 pKa = 10.53RR248 pKa = 11.84GFQRR252 pKa = 11.84SVSTYY257 pKa = 9.88LNTKK261 pKa = 8.96KK262 pKa = 10.23RR263 pKa = 11.84YY264 pKa = 8.89PYY266 pKa = 10.24ISS268 pKa = 3.23
MM1 pKa = 7.36SAYY4 pKa = 9.44TEE6 pKa = 4.22ALQLQRR12 pKa = 11.84DD13 pKa = 3.89AQSFTKK19 pKa = 9.66QQWNFEE25 pKa = 4.33QNLSSTAHH33 pKa = 4.86QRR35 pKa = 11.84EE36 pKa = 4.02VSDD39 pKa = 4.25LKK41 pKa = 11.07KK42 pKa = 10.88AGLNPVLSANSGASTPSVGLDD63 pKa = 3.11SGVQAASGYY72 pKa = 9.06MSSKK76 pKa = 8.64TAAAATRR83 pKa = 11.84YY84 pKa = 9.19ASNMSYY90 pKa = 10.81KK91 pKa = 10.49AALAQASATRR101 pKa = 11.84YY102 pKa = 8.86AASLNYY108 pKa = 8.26EE109 pKa = 4.13AQMSSNKK116 pKa = 10.16YY117 pKa = 9.37GIAMQIANSAAGEE130 pKa = 4.16GGVIGFIKK138 pKa = 10.34RR139 pKa = 11.84AVNSAKK145 pKa = 10.38SYY147 pKa = 11.0NSSIDD152 pKa = 3.41NTVKK156 pKa = 10.77SNNISIPNVKK166 pKa = 8.45VTPQSVLNGKK176 pKa = 8.9ALNGVKK182 pKa = 10.11RR183 pKa = 11.84EE184 pKa = 4.14DD185 pKa = 3.48LLKK188 pKa = 10.46PVTSSSQYY196 pKa = 9.7QFGNKK201 pKa = 6.86VTLNDD206 pKa = 3.32TGKK209 pKa = 10.59KK210 pKa = 9.33YY211 pKa = 10.63VSKK214 pKa = 10.73VLSDD218 pKa = 4.01LGVSTKK224 pKa = 10.87APIGKK229 pKa = 8.78EE230 pKa = 3.82CQIYY234 pKa = 8.88LQQYY238 pKa = 9.52LITGNLVYY246 pKa = 10.82YY247 pKa = 10.53RR248 pKa = 11.84GFQRR252 pKa = 11.84SVSTYY257 pKa = 9.88LNTKK261 pKa = 8.96KK262 pKa = 10.23RR263 pKa = 11.84YY264 pKa = 8.89PYY266 pKa = 10.24ISS268 pKa = 3.23
Molecular weight: 28.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1423 |
80 |
565 |
284.6 |
32.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.833 ± 1.055 | 1.546 ± 0.4 |
6.676 ± 0.828 | 4.076 ± 0.538 |
4.638 ± 0.649 | 6.184 ± 0.236 |
1.476 ± 0.251 | 6.325 ± 0.561 |
5.341 ± 1.304 | 7.238 ± 0.691 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.319 ± 0.486 | 7.449 ± 1.133 |
4.216 ± 0.691 | 3.584 ± 0.631 |
4.989 ± 0.591 | 10.049 ± 1.509 |
5.411 ± 0.335 | 5.13 ± 1.05 |
1.405 ± 0.277 | 6.114 ± 0.998 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
