
Erysiphe necator mitovirus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Lenarviricota; Howeltoviricetes; Cryppavirales; Mitoviridae; Mitovirus; unclassified Mitovirus
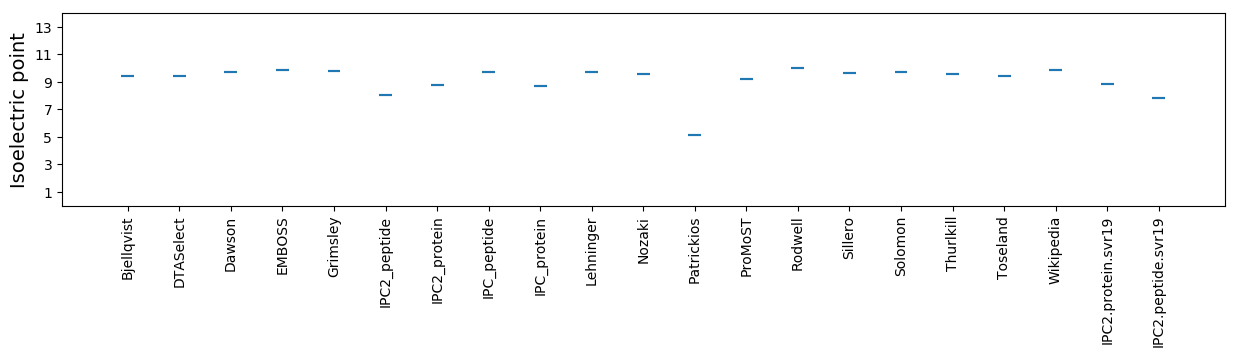
Average proteome isoelectric point is 8.84
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
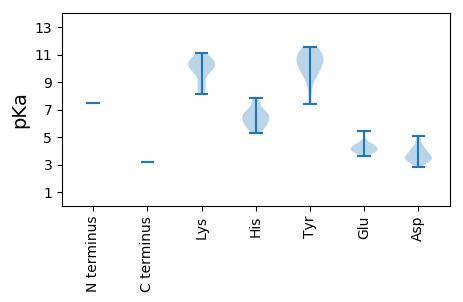
Protein with the lowest isoelectric point:
>tr|A0A2I5ARE9|A0A2I5ARE9_9VIRU RNA-dependent RNA polymerase OS=Erysiphe necator mitovirus 2 OX=2052562 PE=4 SV=1
MM1 pKa = 7.47LWTGRR6 pKa = 11.84SYY8 pKa = 10.9FPSIPEE14 pKa = 3.78EE15 pKa = 3.86LLQEE19 pKa = 4.31SVRR22 pKa = 11.84KK23 pKa = 10.35LEE25 pKa = 4.04TDD27 pKa = 4.15FEE29 pKa = 5.24NNLSRR34 pKa = 11.84GRR36 pKa = 11.84VAVSDD41 pKa = 3.39KK42 pKa = 10.63FKK44 pKa = 10.85AYY46 pKa = 10.14RR47 pKa = 11.84LSLTRR52 pKa = 11.84YY53 pKa = 9.21VSGSPIRR60 pKa = 11.84LEE62 pKa = 5.45GIVQDD67 pKa = 4.22RR68 pKa = 11.84NLLPTVLPVDD78 pKa = 4.16LRR80 pKa = 11.84RR81 pKa = 11.84LIVNRR86 pKa = 11.84DD87 pKa = 3.09LNAIRR92 pKa = 11.84WALTLFTISRR102 pKa = 11.84TVMGGKK108 pKa = 8.74PVNYY112 pKa = 9.77DD113 pKa = 3.6DD114 pKa = 3.87IVRR117 pKa = 11.84PGPHH121 pKa = 7.63PDD123 pKa = 3.48QIHH126 pKa = 5.84TEE128 pKa = 3.64ISDD131 pKa = 3.69YY132 pKa = 11.55DD133 pKa = 3.95VVQFWQRR140 pKa = 11.84LGRR143 pKa = 11.84PKK145 pKa = 10.09IDD147 pKa = 3.3WKK149 pKa = 10.14WKK151 pKa = 8.12NFHH154 pKa = 6.8WSTKK158 pKa = 9.94SGPNGPALQGALADD172 pKa = 4.69LKK174 pKa = 11.09CLTPDD179 pKa = 5.07LIQAIDD185 pKa = 3.14SFMPVKK191 pKa = 10.91NPLTTLLEE199 pKa = 4.57LKK201 pKa = 10.45DD202 pKa = 4.25SPLVAHH208 pKa = 7.85WYY210 pKa = 9.76QVFKK214 pKa = 11.08VKK216 pKa = 10.62SRR218 pKa = 11.84GKK220 pKa = 9.99LRR222 pKa = 11.84KK223 pKa = 9.61LSIKK227 pKa = 10.14DD228 pKa = 3.18DD229 pKa = 3.98RR230 pKa = 11.84EE231 pKa = 3.95AKK233 pKa = 10.32SRR235 pKa = 11.84VFAILDD241 pKa = 3.56YY242 pKa = 10.39WSQTALQEE250 pKa = 4.01LHH252 pKa = 6.78KK253 pKa = 10.96GAFKK257 pKa = 10.92LLRR260 pKa = 11.84RR261 pKa = 11.84LPRR264 pKa = 11.84DD265 pKa = 3.37CTYY268 pKa = 11.43DD269 pKa = 3.28QGKK272 pKa = 9.23LLSEE276 pKa = 4.56LANKK280 pKa = 8.24KK281 pKa = 8.33TDD283 pKa = 3.11HH284 pKa = 6.33QYY286 pKa = 11.46YY287 pKa = 10.67SFDD290 pKa = 3.82LSSATDD296 pKa = 4.02RR297 pKa = 11.84FPLDD301 pKa = 2.99IQEE304 pKa = 4.26RR305 pKa = 11.84FVRR308 pKa = 11.84LLTNKK313 pKa = 9.91DD314 pKa = 3.37VAKK317 pKa = 9.69SWRR320 pKa = 11.84KK321 pKa = 8.79ILVGEE326 pKa = 4.62EE327 pKa = 3.86YY328 pKa = 10.62VAPNGLSYY336 pKa = 10.78KK337 pKa = 9.72YY338 pKa = 10.92GCGQPMGAHH347 pKa = 6.01SSWPIFTLCHH357 pKa = 5.65HH358 pKa = 6.14MVVFVAARR366 pKa = 11.84NCGIKK371 pKa = 10.18RR372 pKa = 11.84FYY374 pKa = 10.95DD375 pKa = 3.71YY376 pKa = 11.19AILGDD381 pKa = 4.99DD382 pKa = 3.61IVISHH387 pKa = 6.87DD388 pKa = 3.55QVAAEE393 pKa = 4.13YY394 pKa = 10.57KK395 pKa = 10.4RR396 pKa = 11.84IISALGVEE404 pKa = 4.3LSEE407 pKa = 4.62AKK409 pKa = 10.36SHH411 pKa = 5.26VSKK414 pKa = 10.34DD415 pKa = 3.07TFEE418 pKa = 4.04FAKK421 pKa = 10.22RR422 pKa = 11.84WFHH425 pKa = 6.55KK426 pKa = 10.44SVEE429 pKa = 3.98ISPFPLHH436 pKa = 6.61GLISTLKK443 pKa = 9.79SWPQLMEE450 pKa = 4.19LLINEE455 pKa = 4.3VPKK458 pKa = 10.57RR459 pKa = 11.84GYY461 pKa = 10.51RR462 pKa = 11.84SILDD466 pKa = 3.61FSPGLNSFVDD476 pKa = 4.34LFPSIRR482 pKa = 11.84MGKK485 pKa = 8.12TILKK489 pKa = 9.98RR490 pKa = 11.84IEE492 pKa = 4.4LNLHH496 pKa = 6.56LPYY499 pKa = 9.76MWTSEE504 pKa = 3.95EE505 pKa = 4.13GDD507 pKa = 3.63VNEE510 pKa = 4.31RR511 pKa = 11.84CQPLFTYY518 pKa = 10.0APSMSHH524 pKa = 6.16WPNDD528 pKa = 3.51QLFSTLTKK536 pKa = 8.86CASMIARR543 pKa = 11.84RR544 pKa = 11.84DD545 pKa = 3.4LGKK548 pKa = 10.56AISDD552 pKa = 3.45VSRR555 pKa = 11.84TLDD558 pKa = 3.82RR559 pKa = 11.84LWSDD563 pKa = 3.52LTSSLEE569 pKa = 4.03SSAVDD574 pKa = 3.27QSASLAPSFVDD585 pKa = 4.09NIPMIKK591 pKa = 9.9VLKK594 pKa = 10.33DD595 pKa = 3.31LSDD598 pKa = 4.76PDD600 pKa = 4.07TQFLGSVPLGKK611 pKa = 8.69LTQGWDD617 pKa = 2.84VWKK620 pKa = 10.15AWRR623 pKa = 11.84NIDD626 pKa = 4.47LLNIPKK632 pKa = 10.49LNGILPMRR640 pKa = 11.84ALEE643 pKa = 3.98RR644 pKa = 11.84KK645 pKa = 8.78VNSRR649 pKa = 11.84AHH651 pKa = 5.63LSLRR655 pKa = 11.84LMQYY659 pKa = 7.43VTKK662 pKa = 10.69LPYY665 pKa = 9.5QQIIEE670 pKa = 4.37EE671 pKa = 4.29LTAYY675 pKa = 8.41DD676 pKa = 3.58APRR679 pKa = 11.84VRR681 pKa = 11.84KK682 pKa = 9.48RR683 pKa = 11.84RR684 pKa = 11.84PRR686 pKa = 11.84SS687 pKa = 3.21
MM1 pKa = 7.47LWTGRR6 pKa = 11.84SYY8 pKa = 10.9FPSIPEE14 pKa = 3.78EE15 pKa = 3.86LLQEE19 pKa = 4.31SVRR22 pKa = 11.84KK23 pKa = 10.35LEE25 pKa = 4.04TDD27 pKa = 4.15FEE29 pKa = 5.24NNLSRR34 pKa = 11.84GRR36 pKa = 11.84VAVSDD41 pKa = 3.39KK42 pKa = 10.63FKK44 pKa = 10.85AYY46 pKa = 10.14RR47 pKa = 11.84LSLTRR52 pKa = 11.84YY53 pKa = 9.21VSGSPIRR60 pKa = 11.84LEE62 pKa = 5.45GIVQDD67 pKa = 4.22RR68 pKa = 11.84NLLPTVLPVDD78 pKa = 4.16LRR80 pKa = 11.84RR81 pKa = 11.84LIVNRR86 pKa = 11.84DD87 pKa = 3.09LNAIRR92 pKa = 11.84WALTLFTISRR102 pKa = 11.84TVMGGKK108 pKa = 8.74PVNYY112 pKa = 9.77DD113 pKa = 3.6DD114 pKa = 3.87IVRR117 pKa = 11.84PGPHH121 pKa = 7.63PDD123 pKa = 3.48QIHH126 pKa = 5.84TEE128 pKa = 3.64ISDD131 pKa = 3.69YY132 pKa = 11.55DD133 pKa = 3.95VVQFWQRR140 pKa = 11.84LGRR143 pKa = 11.84PKK145 pKa = 10.09IDD147 pKa = 3.3WKK149 pKa = 10.14WKK151 pKa = 8.12NFHH154 pKa = 6.8WSTKK158 pKa = 9.94SGPNGPALQGALADD172 pKa = 4.69LKK174 pKa = 11.09CLTPDD179 pKa = 5.07LIQAIDD185 pKa = 3.14SFMPVKK191 pKa = 10.91NPLTTLLEE199 pKa = 4.57LKK201 pKa = 10.45DD202 pKa = 4.25SPLVAHH208 pKa = 7.85WYY210 pKa = 9.76QVFKK214 pKa = 11.08VKK216 pKa = 10.62SRR218 pKa = 11.84GKK220 pKa = 9.99LRR222 pKa = 11.84KK223 pKa = 9.61LSIKK227 pKa = 10.14DD228 pKa = 3.18DD229 pKa = 3.98RR230 pKa = 11.84EE231 pKa = 3.95AKK233 pKa = 10.32SRR235 pKa = 11.84VFAILDD241 pKa = 3.56YY242 pKa = 10.39WSQTALQEE250 pKa = 4.01LHH252 pKa = 6.78KK253 pKa = 10.96GAFKK257 pKa = 10.92LLRR260 pKa = 11.84RR261 pKa = 11.84LPRR264 pKa = 11.84DD265 pKa = 3.37CTYY268 pKa = 11.43DD269 pKa = 3.28QGKK272 pKa = 9.23LLSEE276 pKa = 4.56LANKK280 pKa = 8.24KK281 pKa = 8.33TDD283 pKa = 3.11HH284 pKa = 6.33QYY286 pKa = 11.46YY287 pKa = 10.67SFDD290 pKa = 3.82LSSATDD296 pKa = 4.02RR297 pKa = 11.84FPLDD301 pKa = 2.99IQEE304 pKa = 4.26RR305 pKa = 11.84FVRR308 pKa = 11.84LLTNKK313 pKa = 9.91DD314 pKa = 3.37VAKK317 pKa = 9.69SWRR320 pKa = 11.84KK321 pKa = 8.79ILVGEE326 pKa = 4.62EE327 pKa = 3.86YY328 pKa = 10.62VAPNGLSYY336 pKa = 10.78KK337 pKa = 9.72YY338 pKa = 10.92GCGQPMGAHH347 pKa = 6.01SSWPIFTLCHH357 pKa = 5.65HH358 pKa = 6.14MVVFVAARR366 pKa = 11.84NCGIKK371 pKa = 10.18RR372 pKa = 11.84FYY374 pKa = 10.95DD375 pKa = 3.71YY376 pKa = 11.19AILGDD381 pKa = 4.99DD382 pKa = 3.61IVISHH387 pKa = 6.87DD388 pKa = 3.55QVAAEE393 pKa = 4.13YY394 pKa = 10.57KK395 pKa = 10.4RR396 pKa = 11.84IISALGVEE404 pKa = 4.3LSEE407 pKa = 4.62AKK409 pKa = 10.36SHH411 pKa = 5.26VSKK414 pKa = 10.34DD415 pKa = 3.07TFEE418 pKa = 4.04FAKK421 pKa = 10.22RR422 pKa = 11.84WFHH425 pKa = 6.55KK426 pKa = 10.44SVEE429 pKa = 3.98ISPFPLHH436 pKa = 6.61GLISTLKK443 pKa = 9.79SWPQLMEE450 pKa = 4.19LLINEE455 pKa = 4.3VPKK458 pKa = 10.57RR459 pKa = 11.84GYY461 pKa = 10.51RR462 pKa = 11.84SILDD466 pKa = 3.61FSPGLNSFVDD476 pKa = 4.34LFPSIRR482 pKa = 11.84MGKK485 pKa = 8.12TILKK489 pKa = 9.98RR490 pKa = 11.84IEE492 pKa = 4.4LNLHH496 pKa = 6.56LPYY499 pKa = 9.76MWTSEE504 pKa = 3.95EE505 pKa = 4.13GDD507 pKa = 3.63VNEE510 pKa = 4.31RR511 pKa = 11.84CQPLFTYY518 pKa = 10.0APSMSHH524 pKa = 6.16WPNDD528 pKa = 3.51QLFSTLTKK536 pKa = 8.86CASMIARR543 pKa = 11.84RR544 pKa = 11.84DD545 pKa = 3.4LGKK548 pKa = 10.56AISDD552 pKa = 3.45VSRR555 pKa = 11.84TLDD558 pKa = 3.82RR559 pKa = 11.84LWSDD563 pKa = 3.52LTSSLEE569 pKa = 4.03SSAVDD574 pKa = 3.27QSASLAPSFVDD585 pKa = 4.09NIPMIKK591 pKa = 9.9VLKK594 pKa = 10.33DD595 pKa = 3.31LSDD598 pKa = 4.76PDD600 pKa = 4.07TQFLGSVPLGKK611 pKa = 8.69LTQGWDD617 pKa = 2.84VWKK620 pKa = 10.15AWRR623 pKa = 11.84NIDD626 pKa = 4.47LLNIPKK632 pKa = 10.49LNGILPMRR640 pKa = 11.84ALEE643 pKa = 3.98RR644 pKa = 11.84KK645 pKa = 8.78VNSRR649 pKa = 11.84AHH651 pKa = 5.63LSLRR655 pKa = 11.84LMQYY659 pKa = 7.43VTKK662 pKa = 10.69LPYY665 pKa = 9.5QQIIEE670 pKa = 4.37EE671 pKa = 4.29LTAYY675 pKa = 8.41DD676 pKa = 3.58APRR679 pKa = 11.84VRR681 pKa = 11.84KK682 pKa = 9.48RR683 pKa = 11.84RR684 pKa = 11.84PRR686 pKa = 11.84SS687 pKa = 3.21
Molecular weight: 78.88 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2I5ARE9|A0A2I5ARE9_9VIRU RNA-dependent RNA polymerase OS=Erysiphe necator mitovirus 2 OX=2052562 PE=4 SV=1
MM1 pKa = 7.47LWTGRR6 pKa = 11.84SYY8 pKa = 10.9FPSIPEE14 pKa = 3.78EE15 pKa = 3.86LLQEE19 pKa = 4.31SVRR22 pKa = 11.84KK23 pKa = 10.35LEE25 pKa = 4.04TDD27 pKa = 4.15FEE29 pKa = 5.24NNLSRR34 pKa = 11.84GRR36 pKa = 11.84VAVSDD41 pKa = 3.39KK42 pKa = 10.63FKK44 pKa = 10.85AYY46 pKa = 10.14RR47 pKa = 11.84LSLTRR52 pKa = 11.84YY53 pKa = 9.21VSGSPIRR60 pKa = 11.84LEE62 pKa = 5.45GIVQDD67 pKa = 4.22RR68 pKa = 11.84NLLPTVLPVDD78 pKa = 4.16LRR80 pKa = 11.84RR81 pKa = 11.84LIVNRR86 pKa = 11.84DD87 pKa = 3.09LNAIRR92 pKa = 11.84WALTLFTISRR102 pKa = 11.84TVMGGKK108 pKa = 8.74PVNYY112 pKa = 9.77DD113 pKa = 3.6DD114 pKa = 3.87IVRR117 pKa = 11.84PGPHH121 pKa = 7.63PDD123 pKa = 3.48QIHH126 pKa = 5.84TEE128 pKa = 3.64ISDD131 pKa = 3.69YY132 pKa = 11.55DD133 pKa = 3.95VVQFWQRR140 pKa = 11.84LGRR143 pKa = 11.84PKK145 pKa = 10.09IDD147 pKa = 3.3WKK149 pKa = 10.14WKK151 pKa = 8.12NFHH154 pKa = 6.8WSTKK158 pKa = 9.94SGPNGPALQGALADD172 pKa = 4.69LKK174 pKa = 11.09CLTPDD179 pKa = 5.07LIQAIDD185 pKa = 3.14SFMPVKK191 pKa = 10.91NPLTTLLEE199 pKa = 4.57LKK201 pKa = 10.45DD202 pKa = 4.25SPLVAHH208 pKa = 7.85WYY210 pKa = 9.76QVFKK214 pKa = 11.08VKK216 pKa = 10.62SRR218 pKa = 11.84GKK220 pKa = 9.99LRR222 pKa = 11.84KK223 pKa = 9.61LSIKK227 pKa = 10.14DD228 pKa = 3.18DD229 pKa = 3.98RR230 pKa = 11.84EE231 pKa = 3.95AKK233 pKa = 10.32SRR235 pKa = 11.84VFAILDD241 pKa = 3.56YY242 pKa = 10.39WSQTALQEE250 pKa = 4.01LHH252 pKa = 6.78KK253 pKa = 10.96GAFKK257 pKa = 10.92LLRR260 pKa = 11.84RR261 pKa = 11.84LPRR264 pKa = 11.84DD265 pKa = 3.37CTYY268 pKa = 11.43DD269 pKa = 3.28QGKK272 pKa = 9.23LLSEE276 pKa = 4.56LANKK280 pKa = 8.24KK281 pKa = 8.33TDD283 pKa = 3.11HH284 pKa = 6.33QYY286 pKa = 11.46YY287 pKa = 10.67SFDD290 pKa = 3.82LSSATDD296 pKa = 4.02RR297 pKa = 11.84FPLDD301 pKa = 2.99IQEE304 pKa = 4.26RR305 pKa = 11.84FVRR308 pKa = 11.84LLTNKK313 pKa = 9.91DD314 pKa = 3.37VAKK317 pKa = 9.69SWRR320 pKa = 11.84KK321 pKa = 8.79ILVGEE326 pKa = 4.62EE327 pKa = 3.86YY328 pKa = 10.62VAPNGLSYY336 pKa = 10.78KK337 pKa = 9.72YY338 pKa = 10.92GCGQPMGAHH347 pKa = 6.01SSWPIFTLCHH357 pKa = 5.65HH358 pKa = 6.14MVVFVAARR366 pKa = 11.84NCGIKK371 pKa = 10.18RR372 pKa = 11.84FYY374 pKa = 10.95DD375 pKa = 3.71YY376 pKa = 11.19AILGDD381 pKa = 4.99DD382 pKa = 3.61IVISHH387 pKa = 6.87DD388 pKa = 3.55QVAAEE393 pKa = 4.13YY394 pKa = 10.57KK395 pKa = 10.4RR396 pKa = 11.84IISALGVEE404 pKa = 4.3LSEE407 pKa = 4.62AKK409 pKa = 10.36SHH411 pKa = 5.26VSKK414 pKa = 10.34DD415 pKa = 3.07TFEE418 pKa = 4.04FAKK421 pKa = 10.22RR422 pKa = 11.84WFHH425 pKa = 6.55KK426 pKa = 10.44SVEE429 pKa = 3.98ISPFPLHH436 pKa = 6.61GLISTLKK443 pKa = 9.79SWPQLMEE450 pKa = 4.19LLINEE455 pKa = 4.3VPKK458 pKa = 10.57RR459 pKa = 11.84GYY461 pKa = 10.51RR462 pKa = 11.84SILDD466 pKa = 3.61FSPGLNSFVDD476 pKa = 4.34LFPSIRR482 pKa = 11.84MGKK485 pKa = 8.12TILKK489 pKa = 9.98RR490 pKa = 11.84IEE492 pKa = 4.4LNLHH496 pKa = 6.56LPYY499 pKa = 9.76MWTSEE504 pKa = 3.95EE505 pKa = 4.13GDD507 pKa = 3.63VNEE510 pKa = 4.31RR511 pKa = 11.84CQPLFTYY518 pKa = 10.0APSMSHH524 pKa = 6.16WPNDD528 pKa = 3.51QLFSTLTKK536 pKa = 8.86CASMIARR543 pKa = 11.84RR544 pKa = 11.84DD545 pKa = 3.4LGKK548 pKa = 10.56AISDD552 pKa = 3.45VSRR555 pKa = 11.84TLDD558 pKa = 3.82RR559 pKa = 11.84LWSDD563 pKa = 3.52LTSSLEE569 pKa = 4.03SSAVDD574 pKa = 3.27QSASLAPSFVDD585 pKa = 4.09NIPMIKK591 pKa = 9.9VLKK594 pKa = 10.33DD595 pKa = 3.31LSDD598 pKa = 4.76PDD600 pKa = 4.07TQFLGSVPLGKK611 pKa = 8.69LTQGWDD617 pKa = 2.84VWKK620 pKa = 10.15AWRR623 pKa = 11.84NIDD626 pKa = 4.47LLNIPKK632 pKa = 10.49LNGILPMRR640 pKa = 11.84ALEE643 pKa = 3.98RR644 pKa = 11.84KK645 pKa = 8.78VNSRR649 pKa = 11.84AHH651 pKa = 5.63LSLRR655 pKa = 11.84LMQYY659 pKa = 7.43VTKK662 pKa = 10.69LPYY665 pKa = 9.5QQIIEE670 pKa = 4.37EE671 pKa = 4.29LTAYY675 pKa = 8.41DD676 pKa = 3.58APRR679 pKa = 11.84VRR681 pKa = 11.84KK682 pKa = 9.48RR683 pKa = 11.84RR684 pKa = 11.84PRR686 pKa = 11.84SS687 pKa = 3.21
MM1 pKa = 7.47LWTGRR6 pKa = 11.84SYY8 pKa = 10.9FPSIPEE14 pKa = 3.78EE15 pKa = 3.86LLQEE19 pKa = 4.31SVRR22 pKa = 11.84KK23 pKa = 10.35LEE25 pKa = 4.04TDD27 pKa = 4.15FEE29 pKa = 5.24NNLSRR34 pKa = 11.84GRR36 pKa = 11.84VAVSDD41 pKa = 3.39KK42 pKa = 10.63FKK44 pKa = 10.85AYY46 pKa = 10.14RR47 pKa = 11.84LSLTRR52 pKa = 11.84YY53 pKa = 9.21VSGSPIRR60 pKa = 11.84LEE62 pKa = 5.45GIVQDD67 pKa = 4.22RR68 pKa = 11.84NLLPTVLPVDD78 pKa = 4.16LRR80 pKa = 11.84RR81 pKa = 11.84LIVNRR86 pKa = 11.84DD87 pKa = 3.09LNAIRR92 pKa = 11.84WALTLFTISRR102 pKa = 11.84TVMGGKK108 pKa = 8.74PVNYY112 pKa = 9.77DD113 pKa = 3.6DD114 pKa = 3.87IVRR117 pKa = 11.84PGPHH121 pKa = 7.63PDD123 pKa = 3.48QIHH126 pKa = 5.84TEE128 pKa = 3.64ISDD131 pKa = 3.69YY132 pKa = 11.55DD133 pKa = 3.95VVQFWQRR140 pKa = 11.84LGRR143 pKa = 11.84PKK145 pKa = 10.09IDD147 pKa = 3.3WKK149 pKa = 10.14WKK151 pKa = 8.12NFHH154 pKa = 6.8WSTKK158 pKa = 9.94SGPNGPALQGALADD172 pKa = 4.69LKK174 pKa = 11.09CLTPDD179 pKa = 5.07LIQAIDD185 pKa = 3.14SFMPVKK191 pKa = 10.91NPLTTLLEE199 pKa = 4.57LKK201 pKa = 10.45DD202 pKa = 4.25SPLVAHH208 pKa = 7.85WYY210 pKa = 9.76QVFKK214 pKa = 11.08VKK216 pKa = 10.62SRR218 pKa = 11.84GKK220 pKa = 9.99LRR222 pKa = 11.84KK223 pKa = 9.61LSIKK227 pKa = 10.14DD228 pKa = 3.18DD229 pKa = 3.98RR230 pKa = 11.84EE231 pKa = 3.95AKK233 pKa = 10.32SRR235 pKa = 11.84VFAILDD241 pKa = 3.56YY242 pKa = 10.39WSQTALQEE250 pKa = 4.01LHH252 pKa = 6.78KK253 pKa = 10.96GAFKK257 pKa = 10.92LLRR260 pKa = 11.84RR261 pKa = 11.84LPRR264 pKa = 11.84DD265 pKa = 3.37CTYY268 pKa = 11.43DD269 pKa = 3.28QGKK272 pKa = 9.23LLSEE276 pKa = 4.56LANKK280 pKa = 8.24KK281 pKa = 8.33TDD283 pKa = 3.11HH284 pKa = 6.33QYY286 pKa = 11.46YY287 pKa = 10.67SFDD290 pKa = 3.82LSSATDD296 pKa = 4.02RR297 pKa = 11.84FPLDD301 pKa = 2.99IQEE304 pKa = 4.26RR305 pKa = 11.84FVRR308 pKa = 11.84LLTNKK313 pKa = 9.91DD314 pKa = 3.37VAKK317 pKa = 9.69SWRR320 pKa = 11.84KK321 pKa = 8.79ILVGEE326 pKa = 4.62EE327 pKa = 3.86YY328 pKa = 10.62VAPNGLSYY336 pKa = 10.78KK337 pKa = 9.72YY338 pKa = 10.92GCGQPMGAHH347 pKa = 6.01SSWPIFTLCHH357 pKa = 5.65HH358 pKa = 6.14MVVFVAARR366 pKa = 11.84NCGIKK371 pKa = 10.18RR372 pKa = 11.84FYY374 pKa = 10.95DD375 pKa = 3.71YY376 pKa = 11.19AILGDD381 pKa = 4.99DD382 pKa = 3.61IVISHH387 pKa = 6.87DD388 pKa = 3.55QVAAEE393 pKa = 4.13YY394 pKa = 10.57KK395 pKa = 10.4RR396 pKa = 11.84IISALGVEE404 pKa = 4.3LSEE407 pKa = 4.62AKK409 pKa = 10.36SHH411 pKa = 5.26VSKK414 pKa = 10.34DD415 pKa = 3.07TFEE418 pKa = 4.04FAKK421 pKa = 10.22RR422 pKa = 11.84WFHH425 pKa = 6.55KK426 pKa = 10.44SVEE429 pKa = 3.98ISPFPLHH436 pKa = 6.61GLISTLKK443 pKa = 9.79SWPQLMEE450 pKa = 4.19LLINEE455 pKa = 4.3VPKK458 pKa = 10.57RR459 pKa = 11.84GYY461 pKa = 10.51RR462 pKa = 11.84SILDD466 pKa = 3.61FSPGLNSFVDD476 pKa = 4.34LFPSIRR482 pKa = 11.84MGKK485 pKa = 8.12TILKK489 pKa = 9.98RR490 pKa = 11.84IEE492 pKa = 4.4LNLHH496 pKa = 6.56LPYY499 pKa = 9.76MWTSEE504 pKa = 3.95EE505 pKa = 4.13GDD507 pKa = 3.63VNEE510 pKa = 4.31RR511 pKa = 11.84CQPLFTYY518 pKa = 10.0APSMSHH524 pKa = 6.16WPNDD528 pKa = 3.51QLFSTLTKK536 pKa = 8.86CASMIARR543 pKa = 11.84RR544 pKa = 11.84DD545 pKa = 3.4LGKK548 pKa = 10.56AISDD552 pKa = 3.45VSRR555 pKa = 11.84TLDD558 pKa = 3.82RR559 pKa = 11.84LWSDD563 pKa = 3.52LTSSLEE569 pKa = 4.03SSAVDD574 pKa = 3.27QSASLAPSFVDD585 pKa = 4.09NIPMIKK591 pKa = 9.9VLKK594 pKa = 10.33DD595 pKa = 3.31LSDD598 pKa = 4.76PDD600 pKa = 4.07TQFLGSVPLGKK611 pKa = 8.69LTQGWDD617 pKa = 2.84VWKK620 pKa = 10.15AWRR623 pKa = 11.84NIDD626 pKa = 4.47LLNIPKK632 pKa = 10.49LNGILPMRR640 pKa = 11.84ALEE643 pKa = 3.98RR644 pKa = 11.84KK645 pKa = 8.78VNSRR649 pKa = 11.84AHH651 pKa = 5.63LSLRR655 pKa = 11.84LMQYY659 pKa = 7.43VTKK662 pKa = 10.69LPYY665 pKa = 9.5QQIIEE670 pKa = 4.37EE671 pKa = 4.29LTAYY675 pKa = 8.41DD676 pKa = 3.58APRR679 pKa = 11.84VRR681 pKa = 11.84KK682 pKa = 9.48RR683 pKa = 11.84RR684 pKa = 11.84PRR686 pKa = 11.84SS687 pKa = 3.21
Molecular weight: 78.88 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
687 |
687 |
687 |
687.0 |
78.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.531 ± 0.0 | 1.019 ± 0.0 |
6.696 ± 0.0 | 4.221 ± 0.0 |
3.93 ± 0.0 | 4.658 ± 0.0 |
2.329 ± 0.0 | 5.677 ± 0.0 |
6.696 ± 0.0 | 12.373 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.892 ± 0.0 | 3.348 ± 0.0 |
5.822 ± 0.0 | 3.493 ± 0.0 |
7.278 ± 0.0 | 8.588 ± 0.0 |
4.512 ± 0.0 | 6.114 ± 0.0 |
2.62 ± 0.0 | 3.202 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
