
Bovine parvovirus (isolate pAT153/1986) (BPV)
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Quintoviricetes; Piccovirales; Parvoviridae; Parvovirinae; Bocaparvovirus; Ungulate bocaparvovirus 1; Bovine parvovirus
Average proteome isoelectric point is 7.64
Get precalculated fractions of proteins
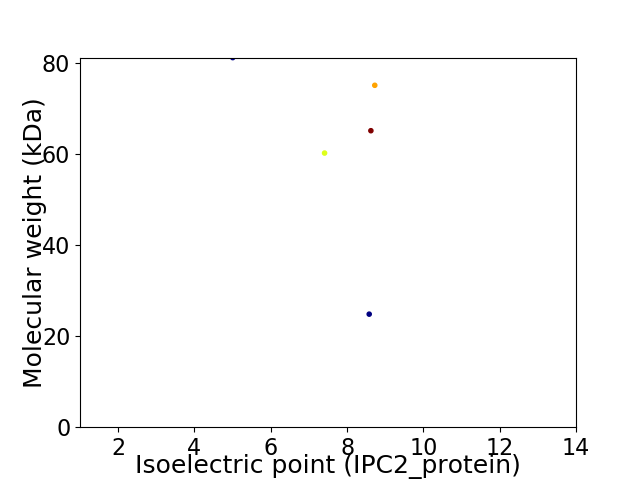
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P07297|CAPSD_PAVBP Minor capsid protein VP1 OS=Bovine parvovirus (isolate pAT153/1986) OX=648243 PE=3 SV=1
MM1 pKa = 6.89QPSRR5 pKa = 11.84GLYY8 pKa = 9.71SALHH12 pKa = 5.58SFKK15 pKa = 10.77HH16 pKa = 5.22LLITIRR22 pKa = 11.84QRR24 pKa = 11.84THH26 pKa = 6.98APDD29 pKa = 3.45TVLSRR34 pKa = 11.84DD35 pKa = 4.34EE36 pKa = 4.08LNAWNWLQKK45 pKa = 9.56IAQRR49 pKa = 11.84VMRR52 pKa = 11.84GDD54 pKa = 3.15MDD56 pKa = 5.98DD57 pKa = 3.64IVDD60 pKa = 3.5ILKK63 pKa = 10.22CRR65 pKa = 11.84KK66 pKa = 10.02ANGTLVAQAINGTEE80 pKa = 4.63FITRR84 pKa = 11.84YY85 pKa = 8.38MLPKK89 pKa = 9.43NRR91 pKa = 11.84KK92 pKa = 8.5VADD95 pKa = 3.77TVLTRR100 pKa = 11.84HH101 pKa = 5.27TTPEE105 pKa = 3.59QSYY108 pKa = 10.86DD109 pKa = 3.74STWGKK114 pKa = 8.49TYY116 pKa = 10.84GFAVCNGEE124 pKa = 4.31TVSEE128 pKa = 4.47FTRR131 pKa = 11.84KK132 pKa = 10.1DD133 pKa = 3.1LWKK136 pKa = 10.37VLYY139 pKa = 10.31NIYY142 pKa = 8.86TAHH145 pKa = 7.21PAEE148 pKa = 4.63NMLNSNPSVWGDD160 pKa = 3.46LPRR163 pKa = 11.84VSANRR168 pKa = 11.84IDD170 pKa = 4.9ADD172 pKa = 3.7DD173 pKa = 4.16AEE175 pKa = 4.56ARR177 pKa = 11.84SRR179 pKa = 11.84PIKK182 pKa = 10.36LSRR185 pKa = 11.84KK186 pKa = 7.07QKK188 pKa = 10.21IMAEE192 pKa = 4.37VIQRR196 pKa = 11.84ATDD199 pKa = 3.86GLLLTYY205 pKa = 10.75NDD207 pKa = 4.26LVVHH211 pKa = 6.58LSDD214 pKa = 5.14LMLMLEE220 pKa = 4.41GMPGGSKK227 pKa = 8.51TAEE230 pKa = 3.89QLLTMIHH237 pKa = 6.5IKK239 pKa = 10.67LCAKK243 pKa = 9.28YY244 pKa = 9.96NAYY247 pKa = 10.35EE248 pKa = 4.04FMLMKK253 pKa = 10.49TPATQNMNPGAPHH266 pKa = 6.78YY267 pKa = 9.83DD268 pKa = 3.48CQGNLVFKK276 pKa = 10.69LLNLQGYY283 pKa = 8.04NPWQVGHH290 pKa = 6.95WLVMMLSKK298 pKa = 10.08KK299 pKa = 7.96TGKK302 pKa = 10.21RR303 pKa = 11.84NSTLFYY309 pKa = 10.93GPASTGKK316 pKa = 8.58TNLAKK321 pKa = 10.49AICHH325 pKa = 5.71AVGLYY330 pKa = 10.87GCVNHH335 pKa = 6.51NNKK338 pKa = 9.06QFPFNDD344 pKa = 3.97APNKK348 pKa = 9.42MILWWEE354 pKa = 4.1EE355 pKa = 4.01CIMTTDD361 pKa = 3.4YY362 pKa = 11.6VEE364 pKa = 4.07AAKK367 pKa = 10.54CVLGGTHH374 pKa = 5.44VRR376 pKa = 11.84VDD378 pKa = 3.75VKK380 pKa = 11.08HH381 pKa = 6.37KK382 pKa = 10.55DD383 pKa = 3.54SRR385 pKa = 11.84EE386 pKa = 3.94LPQIPVLLSSNHH398 pKa = 6.23DD399 pKa = 3.55VYY401 pKa = 11.37TVVGGNATFGVHH413 pKa = 6.97AAPLKK418 pKa = 10.4EE419 pKa = 5.63RR420 pKa = 11.84ITQMNFMKK428 pKa = 10.37QLPNTFGEE436 pKa = 4.44ITPGMISNWLSHH448 pKa = 6.66CAHH451 pKa = 7.26IHH453 pKa = 5.0QEE455 pKa = 4.07HH456 pKa = 6.83LSLEE460 pKa = 4.18GFAIKK465 pKa = 9.86WDD467 pKa = 3.79VQSVGNSFPLQTLCPGHH484 pKa = 5.34SQNWTFSEE492 pKa = 5.01NGVCWHH498 pKa = 7.05CGGFIQPTPEE508 pKa = 4.33SDD510 pKa = 3.16TDD512 pKa = 3.75SDD514 pKa = 4.72GDD516 pKa = 4.16PDD518 pKa = 4.31PDD520 pKa = 3.69GAVAGDD526 pKa = 4.07SDD528 pKa = 3.77TSANSEE534 pKa = 4.31STVSFSSNDD543 pKa = 3.3SGLGSVTSSAPSVPDD558 pKa = 3.12RR559 pKa = 11.84AEE561 pKa = 4.22EE562 pKa = 3.96IEE564 pKa = 4.71EE565 pKa = 4.16IPSEE569 pKa = 4.03CLEE572 pKa = 4.06WMRR575 pKa = 11.84EE576 pKa = 3.98EE577 pKa = 4.12VDD579 pKa = 3.44RR580 pKa = 11.84LSAHH584 pKa = 7.39DD585 pKa = 4.12INSLAHH591 pKa = 5.56QATGFILDD599 pKa = 5.39PIPEE603 pKa = 4.14EE604 pKa = 4.24PEE606 pKa = 3.56EE607 pKa = 4.41GEE609 pKa = 3.77RR610 pKa = 11.84DD611 pKa = 3.59LARR614 pKa = 11.84EE615 pKa = 3.95DD616 pKa = 4.35AEE618 pKa = 4.58PEE620 pKa = 3.82ASTSHH625 pKa = 5.79TPATKK630 pKa = 9.76RR631 pKa = 11.84ARR633 pKa = 11.84VEE635 pKa = 4.17EE636 pKa = 4.39GEE638 pKa = 4.35PWDD641 pKa = 3.56GTQPITEE648 pKa = 4.47GDD650 pKa = 3.55WIDD653 pKa = 3.47FEE655 pKa = 5.14SRR657 pKa = 11.84QKK659 pKa = 10.42RR660 pKa = 11.84RR661 pKa = 11.84RR662 pKa = 11.84LEE664 pKa = 3.87RR665 pKa = 11.84EE666 pKa = 3.73EE667 pKa = 4.89KK668 pKa = 10.7GGEE671 pKa = 4.05DD672 pKa = 3.4EE673 pKa = 5.37DD674 pKa = 4.81MEE676 pKa = 4.51VQEE679 pKa = 5.19SDD681 pKa = 3.04PSAWGEE687 pKa = 3.75KK688 pKa = 10.23LGIVEE693 pKa = 4.81KK694 pKa = 10.44PGEE697 pKa = 4.27EE698 pKa = 4.34PIVLYY703 pKa = 10.73CFEE706 pKa = 4.77TLPEE710 pKa = 4.22SDD712 pKa = 4.77EE713 pKa = 4.77EE714 pKa = 5.01GDD716 pKa = 3.54SDD718 pKa = 5.57KK719 pKa = 11.35EE720 pKa = 4.43NKK722 pKa = 7.8THH724 pKa = 5.93TVV726 pKa = 3.01
MM1 pKa = 6.89QPSRR5 pKa = 11.84GLYY8 pKa = 9.71SALHH12 pKa = 5.58SFKK15 pKa = 10.77HH16 pKa = 5.22LLITIRR22 pKa = 11.84QRR24 pKa = 11.84THH26 pKa = 6.98APDD29 pKa = 3.45TVLSRR34 pKa = 11.84DD35 pKa = 4.34EE36 pKa = 4.08LNAWNWLQKK45 pKa = 9.56IAQRR49 pKa = 11.84VMRR52 pKa = 11.84GDD54 pKa = 3.15MDD56 pKa = 5.98DD57 pKa = 3.64IVDD60 pKa = 3.5ILKK63 pKa = 10.22CRR65 pKa = 11.84KK66 pKa = 10.02ANGTLVAQAINGTEE80 pKa = 4.63FITRR84 pKa = 11.84YY85 pKa = 8.38MLPKK89 pKa = 9.43NRR91 pKa = 11.84KK92 pKa = 8.5VADD95 pKa = 3.77TVLTRR100 pKa = 11.84HH101 pKa = 5.27TTPEE105 pKa = 3.59QSYY108 pKa = 10.86DD109 pKa = 3.74STWGKK114 pKa = 8.49TYY116 pKa = 10.84GFAVCNGEE124 pKa = 4.31TVSEE128 pKa = 4.47FTRR131 pKa = 11.84KK132 pKa = 10.1DD133 pKa = 3.1LWKK136 pKa = 10.37VLYY139 pKa = 10.31NIYY142 pKa = 8.86TAHH145 pKa = 7.21PAEE148 pKa = 4.63NMLNSNPSVWGDD160 pKa = 3.46LPRR163 pKa = 11.84VSANRR168 pKa = 11.84IDD170 pKa = 4.9ADD172 pKa = 3.7DD173 pKa = 4.16AEE175 pKa = 4.56ARR177 pKa = 11.84SRR179 pKa = 11.84PIKK182 pKa = 10.36LSRR185 pKa = 11.84KK186 pKa = 7.07QKK188 pKa = 10.21IMAEE192 pKa = 4.37VIQRR196 pKa = 11.84ATDD199 pKa = 3.86GLLLTYY205 pKa = 10.75NDD207 pKa = 4.26LVVHH211 pKa = 6.58LSDD214 pKa = 5.14LMLMLEE220 pKa = 4.41GMPGGSKK227 pKa = 8.51TAEE230 pKa = 3.89QLLTMIHH237 pKa = 6.5IKK239 pKa = 10.67LCAKK243 pKa = 9.28YY244 pKa = 9.96NAYY247 pKa = 10.35EE248 pKa = 4.04FMLMKK253 pKa = 10.49TPATQNMNPGAPHH266 pKa = 6.78YY267 pKa = 9.83DD268 pKa = 3.48CQGNLVFKK276 pKa = 10.69LLNLQGYY283 pKa = 8.04NPWQVGHH290 pKa = 6.95WLVMMLSKK298 pKa = 10.08KK299 pKa = 7.96TGKK302 pKa = 10.21RR303 pKa = 11.84NSTLFYY309 pKa = 10.93GPASTGKK316 pKa = 8.58TNLAKK321 pKa = 10.49AICHH325 pKa = 5.71AVGLYY330 pKa = 10.87GCVNHH335 pKa = 6.51NNKK338 pKa = 9.06QFPFNDD344 pKa = 3.97APNKK348 pKa = 9.42MILWWEE354 pKa = 4.1EE355 pKa = 4.01CIMTTDD361 pKa = 3.4YY362 pKa = 11.6VEE364 pKa = 4.07AAKK367 pKa = 10.54CVLGGTHH374 pKa = 5.44VRR376 pKa = 11.84VDD378 pKa = 3.75VKK380 pKa = 11.08HH381 pKa = 6.37KK382 pKa = 10.55DD383 pKa = 3.54SRR385 pKa = 11.84EE386 pKa = 3.94LPQIPVLLSSNHH398 pKa = 6.23DD399 pKa = 3.55VYY401 pKa = 11.37TVVGGNATFGVHH413 pKa = 6.97AAPLKK418 pKa = 10.4EE419 pKa = 5.63RR420 pKa = 11.84ITQMNFMKK428 pKa = 10.37QLPNTFGEE436 pKa = 4.44ITPGMISNWLSHH448 pKa = 6.66CAHH451 pKa = 7.26IHH453 pKa = 5.0QEE455 pKa = 4.07HH456 pKa = 6.83LSLEE460 pKa = 4.18GFAIKK465 pKa = 9.86WDD467 pKa = 3.79VQSVGNSFPLQTLCPGHH484 pKa = 5.34SQNWTFSEE492 pKa = 5.01NGVCWHH498 pKa = 7.05CGGFIQPTPEE508 pKa = 4.33SDD510 pKa = 3.16TDD512 pKa = 3.75SDD514 pKa = 4.72GDD516 pKa = 4.16PDD518 pKa = 4.31PDD520 pKa = 3.69GAVAGDD526 pKa = 4.07SDD528 pKa = 3.77TSANSEE534 pKa = 4.31STVSFSSNDD543 pKa = 3.3SGLGSVTSSAPSVPDD558 pKa = 3.12RR559 pKa = 11.84AEE561 pKa = 4.22EE562 pKa = 3.96IEE564 pKa = 4.71EE565 pKa = 4.16IPSEE569 pKa = 4.03CLEE572 pKa = 4.06WMRR575 pKa = 11.84EE576 pKa = 3.98EE577 pKa = 4.12VDD579 pKa = 3.44RR580 pKa = 11.84LSAHH584 pKa = 7.39DD585 pKa = 4.12INSLAHH591 pKa = 5.56QATGFILDD599 pKa = 5.39PIPEE603 pKa = 4.14EE604 pKa = 4.24PEE606 pKa = 3.56EE607 pKa = 4.41GEE609 pKa = 3.77RR610 pKa = 11.84DD611 pKa = 3.59LARR614 pKa = 11.84EE615 pKa = 3.95DD616 pKa = 4.35AEE618 pKa = 4.58PEE620 pKa = 3.82ASTSHH625 pKa = 5.79TPATKK630 pKa = 9.76RR631 pKa = 11.84ARR633 pKa = 11.84VEE635 pKa = 4.17EE636 pKa = 4.39GEE638 pKa = 4.35PWDD641 pKa = 3.56GTQPITEE648 pKa = 4.47GDD650 pKa = 3.55WIDD653 pKa = 3.47FEE655 pKa = 5.14SRR657 pKa = 11.84QKK659 pKa = 10.42RR660 pKa = 11.84RR661 pKa = 11.84RR662 pKa = 11.84LEE664 pKa = 3.87RR665 pKa = 11.84EE666 pKa = 3.73EE667 pKa = 4.89KK668 pKa = 10.7GGEE671 pKa = 4.05DD672 pKa = 3.4EE673 pKa = 5.37DD674 pKa = 4.81MEE676 pKa = 4.51VQEE679 pKa = 5.19SDD681 pKa = 3.04PSAWGEE687 pKa = 3.75KK688 pKa = 10.23LGIVEE693 pKa = 4.81KK694 pKa = 10.44PGEE697 pKa = 4.27EE698 pKa = 4.34PIVLYY703 pKa = 10.73CFEE706 pKa = 4.77TLPEE710 pKa = 4.22SDD712 pKa = 4.77EE713 pKa = 4.77EE714 pKa = 5.01GDD716 pKa = 3.54SDD718 pKa = 5.57KK719 pKa = 11.35EE720 pKa = 4.43NKK722 pKa = 7.8THH724 pKa = 5.93TVV726 pKa = 3.01
Molecular weight: 81.19 kDa
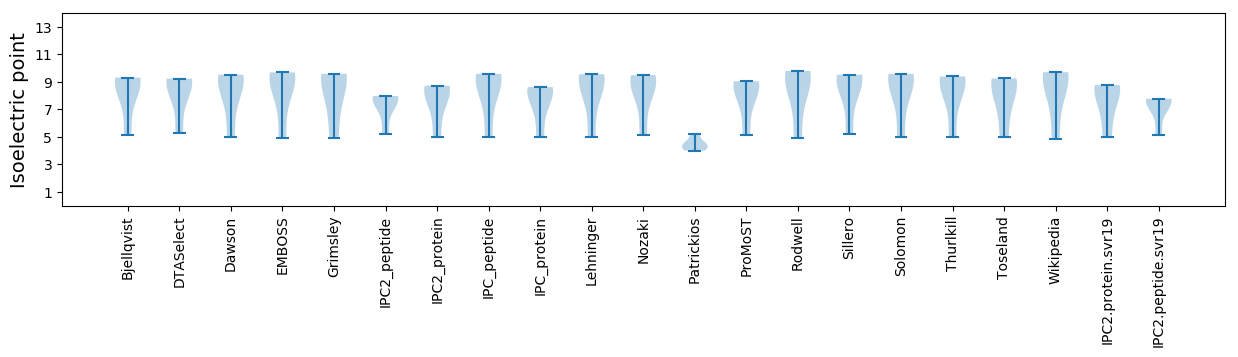
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P07296|NS1_PAVBP Initiator protein NS1 OS=Bovine parvovirus (isolate pAT153/1986) OX=648243 GN=NS1 PE=3 SV=1
MM1 pKa = 7.29EE2 pKa = 5.88RR3 pKa = 11.84SRR5 pKa = 11.84SPRR8 pKa = 11.84EE9 pKa = 3.72TGSTSSRR16 pKa = 11.84DD17 pKa = 3.31KK18 pKa = 11.47SDD20 pKa = 4.18ADD22 pKa = 2.87WSEE25 pKa = 3.52RR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84EE29 pKa = 3.79EE30 pKa = 4.0RR31 pKa = 11.84TRR33 pKa = 11.84TWKK36 pKa = 10.31SRR38 pKa = 11.84SPIRR42 pKa = 11.84ARR44 pKa = 11.84GEE46 pKa = 3.99RR47 pKa = 11.84SWGSWRR53 pKa = 11.84SRR55 pKa = 11.84EE56 pKa = 4.13KK57 pKa = 10.8NQSSSTASRR66 pKa = 11.84PYY68 pKa = 10.36QKK70 pKa = 9.82ATRR73 pKa = 11.84KK74 pKa = 7.55EE75 pKa = 3.98TATKK79 pKa = 8.43KK80 pKa = 8.76TKK82 pKa = 8.68HH83 pKa = 5.3TPFNVFSAHH92 pKa = 6.68RR93 pKa = 11.84ALSKK97 pKa = 10.07TDD99 pKa = 3.37LQFCGFYY106 pKa = 9.43WHH108 pKa = 5.99STRR111 pKa = 11.84LASKK115 pKa = 8.4GTNEE119 pKa = 4.09IFNGLKK125 pKa = 10.17QSFQSKK131 pKa = 10.59AIDD134 pKa = 4.28GKK136 pKa = 11.05LDD138 pKa = 3.34WEE140 pKa = 4.96GVRR143 pKa = 11.84EE144 pKa = 4.4LLFEE148 pKa = 4.28QKK150 pKa = 10.29KK151 pKa = 10.76CLDD154 pKa = 2.83TWYY157 pKa = 10.68RR158 pKa = 11.84NMMYY162 pKa = 10.22HH163 pKa = 6.52FALGGDD169 pKa = 4.11CEE171 pKa = 4.37KK172 pKa = 11.01CNYY175 pKa = 8.82WDD177 pKa = 4.26DD178 pKa = 3.88VYY180 pKa = 11.36KK181 pKa = 10.92KK182 pKa = 9.99HH183 pKa = 6.29LANVDD188 pKa = 3.67TYY190 pKa = 11.65SVAEE194 pKa = 4.66EE195 pKa = 4.08ITDD198 pKa = 4.2SEE200 pKa = 4.27MLEE203 pKa = 3.92AAEE206 pKa = 5.29AVDD209 pKa = 5.04AANQQ213 pKa = 3.32
MM1 pKa = 7.29EE2 pKa = 5.88RR3 pKa = 11.84SRR5 pKa = 11.84SPRR8 pKa = 11.84EE9 pKa = 3.72TGSTSSRR16 pKa = 11.84DD17 pKa = 3.31KK18 pKa = 11.47SDD20 pKa = 4.18ADD22 pKa = 2.87WSEE25 pKa = 3.52RR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84EE29 pKa = 3.79EE30 pKa = 4.0RR31 pKa = 11.84TRR33 pKa = 11.84TWKK36 pKa = 10.31SRR38 pKa = 11.84SPIRR42 pKa = 11.84ARR44 pKa = 11.84GEE46 pKa = 3.99RR47 pKa = 11.84SWGSWRR53 pKa = 11.84SRR55 pKa = 11.84EE56 pKa = 4.13KK57 pKa = 10.8NQSSSTASRR66 pKa = 11.84PYY68 pKa = 10.36QKK70 pKa = 9.82ATRR73 pKa = 11.84KK74 pKa = 7.55EE75 pKa = 3.98TATKK79 pKa = 8.43KK80 pKa = 8.76TKK82 pKa = 8.68HH83 pKa = 5.3TPFNVFSAHH92 pKa = 6.68RR93 pKa = 11.84ALSKK97 pKa = 10.07TDD99 pKa = 3.37LQFCGFYY106 pKa = 9.43WHH108 pKa = 5.99STRR111 pKa = 11.84LASKK115 pKa = 8.4GTNEE119 pKa = 4.09IFNGLKK125 pKa = 10.17QSFQSKK131 pKa = 10.59AIDD134 pKa = 4.28GKK136 pKa = 11.05LDD138 pKa = 3.34WEE140 pKa = 4.96GVRR143 pKa = 11.84EE144 pKa = 4.4LLFEE148 pKa = 4.28QKK150 pKa = 10.29KK151 pKa = 10.76CLDD154 pKa = 2.83TWYY157 pKa = 10.68RR158 pKa = 11.84NMMYY162 pKa = 10.22HH163 pKa = 6.52FALGGDD169 pKa = 4.11CEE171 pKa = 4.37KK172 pKa = 11.01CNYY175 pKa = 8.82WDD177 pKa = 4.26DD178 pKa = 3.88VYY180 pKa = 11.36KK181 pKa = 10.92KK182 pKa = 9.99HH183 pKa = 6.29LANVDD188 pKa = 3.67TYY190 pKa = 11.65SVAEE194 pKa = 4.66EE195 pKa = 4.08ITDD198 pKa = 4.2SEE200 pKa = 4.27MLEE203 pKa = 3.92AAEE206 pKa = 5.29AVDD209 pKa = 5.04AANQQ213 pKa = 3.32
Molecular weight: 24.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
2730 |
213 |
726 |
546.0 |
61.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.85 ± 0.208 | 1.099 ± 0.275 |
5.604 ± 0.274 | 5.275 ± 0.973 |
2.637 ± 0.177 | 7.766 ± 0.486 |
3.15 ± 0.128 | 4.835 ± 0.389 |
4.945 ± 0.492 | 6.63 ± 0.471 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.784 ± 0.144 | 6.63 ± 0.611 |
5.238 ± 0.422 | 4.835 ± 0.441 |
5.861 ± 0.577 | 6.923 ± 0.532 |
6.74 ± 0.128 | 6.117 ± 0.444 |
2.344 ± 0.194 | 3.736 ± 0.461 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
