
Paracraurococcus sp. NE82
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Acetobacteraceae; Paracraurococcus; unclassified Paracraurococcus
Average proteome isoelectric point is 7.08
Get precalculated fractions of proteins
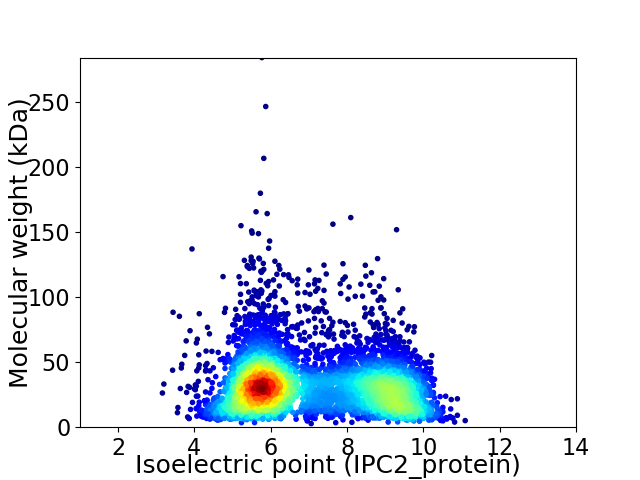
Virtual 2D-PAGE plot for 5359 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4R4D8A5|A0A4R4D8A5_9PROT Cytochrome P450 OS=Paracraurococcus sp. NE82 OX=2527868 GN=EXY23_19725 PE=3 SV=1
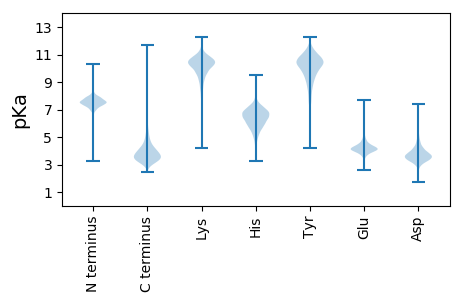
MM1 pKa = 7.76IYY3 pKa = 10.67LNAVNSSLSTSASAVKK19 pKa = 9.59WLATSGTGSTLLGTTLNDD37 pKa = 3.51YY38 pKa = 11.04LSTSDD43 pKa = 3.45PTATLGGGKK52 pKa = 10.2GDD54 pKa = 3.4DD55 pKa = 3.52WYY57 pKa = 11.04AVSDD61 pKa = 3.8YY62 pKa = 9.84RR63 pKa = 11.84TTILEE68 pKa = 4.29TTGNGIDD75 pKa = 4.31TVQSWASRR83 pKa = 11.84FTLPAFVEE91 pKa = 4.01NLQLVGSNQLGTGNALNNIILAKK114 pKa = 9.63TGNATLDD121 pKa = 3.46GGAGNDD127 pKa = 3.48VLVGGTGADD136 pKa = 3.5TFVMSQGNGSDD147 pKa = 3.89AIYY150 pKa = 10.68SFNAAQDD157 pKa = 3.91TIKK160 pKa = 10.88LQGYY164 pKa = 7.07GQYY167 pKa = 11.14GFAAVKK173 pKa = 10.64AGMTQVGADD182 pKa = 3.47TVIALGGGEE191 pKa = 4.52KK192 pKa = 10.42LVLKK196 pKa = 10.33NVQASSLTAQNFSLPSDD213 pKa = 3.59PAHH216 pKa = 7.09FGRR219 pKa = 11.84AMTFGDD225 pKa = 4.01EE226 pKa = 4.25FDD228 pKa = 4.17SLSASGSGLGTDD240 pKa = 4.63WKK242 pKa = 7.78TTWKK246 pKa = 10.28IGVQGRR252 pKa = 11.84TFGTASDD259 pKa = 3.36QAYY262 pKa = 10.67YY263 pKa = 11.25GDD265 pKa = 4.83ASTGTNPFSIEE276 pKa = 4.08DD277 pKa = 4.47GILGITASPGGAAGMGYY294 pKa = 8.81TSGVLTTAKK303 pKa = 10.39SFAQLYY309 pKa = 10.33GYY311 pKa = 9.77FEE313 pKa = 5.09ISAQMPVGEE322 pKa = 4.95GYY324 pKa = 10.88LPAFWLLPTTGAWPPEE340 pKa = 3.64IDD342 pKa = 3.0IFEE345 pKa = 4.41YY346 pKa = 10.36VAKK349 pKa = 10.34DD350 pKa = 3.08PTAIYY355 pKa = 9.52TSYY358 pKa = 9.59GTTAGGATYY367 pKa = 8.39RR368 pKa = 11.84TAHH371 pKa = 6.63LSDD374 pKa = 4.36LSSGFHH380 pKa = 6.47TYY382 pKa = 10.27GLSWQADD389 pKa = 3.54LMKK392 pKa = 10.03WYY394 pKa = 10.94VDD396 pKa = 3.4DD397 pKa = 3.83TLVYY401 pKa = 10.09QVATPAEE408 pKa = 4.7LKK410 pKa = 10.34QPMYY414 pKa = 10.17MLVSLLTGTANSWQGLPTAANPTGTLQVDD443 pKa = 3.56YY444 pKa = 11.2VRR446 pKa = 11.84AYY448 pKa = 10.48QSTAVNVDD456 pKa = 3.32DD457 pKa = 5.53AGDD460 pKa = 3.45VAALGGHH467 pKa = 5.08YY468 pKa = 8.84TRR470 pKa = 11.84NADD473 pKa = 4.37GSDD476 pKa = 3.67LYY478 pKa = 11.44DD479 pKa = 3.78FTDD482 pKa = 3.54SAAMLVMDD490 pKa = 5.46ASGLSSGLHH499 pKa = 5.34GVLAGAAGSQVRR511 pKa = 11.84GSAGPMSFTGGAGVDD526 pKa = 3.43SFFFGVGNATVSGGDD541 pKa = 3.62GNDD544 pKa = 3.19SFVLTRR550 pKa = 11.84GVIAAGDD557 pKa = 3.89VIGDD561 pKa = 3.54FHH563 pKa = 8.55LDD565 pKa = 3.29LGNGTEE571 pKa = 4.27HH572 pKa = 7.5DD573 pKa = 3.97QLQLAGFSGDD583 pKa = 2.95AHH585 pKa = 7.76LDD587 pKa = 3.59VVSASGSTQIYY598 pKa = 9.08RR599 pKa = 11.84VVDD602 pKa = 3.66GTYY605 pKa = 9.86VSPNITIAVVNGAAHH620 pKa = 7.35LSAADD625 pKa = 3.6YY626 pKa = 11.57AFVV629 pKa = 3.54
MM1 pKa = 7.76IYY3 pKa = 10.67LNAVNSSLSTSASAVKK19 pKa = 9.59WLATSGTGSTLLGTTLNDD37 pKa = 3.51YY38 pKa = 11.04LSTSDD43 pKa = 3.45PTATLGGGKK52 pKa = 10.2GDD54 pKa = 3.4DD55 pKa = 3.52WYY57 pKa = 11.04AVSDD61 pKa = 3.8YY62 pKa = 9.84RR63 pKa = 11.84TTILEE68 pKa = 4.29TTGNGIDD75 pKa = 4.31TVQSWASRR83 pKa = 11.84FTLPAFVEE91 pKa = 4.01NLQLVGSNQLGTGNALNNIILAKK114 pKa = 9.63TGNATLDD121 pKa = 3.46GGAGNDD127 pKa = 3.48VLVGGTGADD136 pKa = 3.5TFVMSQGNGSDD147 pKa = 3.89AIYY150 pKa = 10.68SFNAAQDD157 pKa = 3.91TIKK160 pKa = 10.88LQGYY164 pKa = 7.07GQYY167 pKa = 11.14GFAAVKK173 pKa = 10.64AGMTQVGADD182 pKa = 3.47TVIALGGGEE191 pKa = 4.52KK192 pKa = 10.42LVLKK196 pKa = 10.33NVQASSLTAQNFSLPSDD213 pKa = 3.59PAHH216 pKa = 7.09FGRR219 pKa = 11.84AMTFGDD225 pKa = 4.01EE226 pKa = 4.25FDD228 pKa = 4.17SLSASGSGLGTDD240 pKa = 4.63WKK242 pKa = 7.78TTWKK246 pKa = 10.28IGVQGRR252 pKa = 11.84TFGTASDD259 pKa = 3.36QAYY262 pKa = 10.67YY263 pKa = 11.25GDD265 pKa = 4.83ASTGTNPFSIEE276 pKa = 4.08DD277 pKa = 4.47GILGITASPGGAAGMGYY294 pKa = 8.81TSGVLTTAKK303 pKa = 10.39SFAQLYY309 pKa = 10.33GYY311 pKa = 9.77FEE313 pKa = 5.09ISAQMPVGEE322 pKa = 4.95GYY324 pKa = 10.88LPAFWLLPTTGAWPPEE340 pKa = 3.64IDD342 pKa = 3.0IFEE345 pKa = 4.41YY346 pKa = 10.36VAKK349 pKa = 10.34DD350 pKa = 3.08PTAIYY355 pKa = 9.52TSYY358 pKa = 9.59GTTAGGATYY367 pKa = 8.39RR368 pKa = 11.84TAHH371 pKa = 6.63LSDD374 pKa = 4.36LSSGFHH380 pKa = 6.47TYY382 pKa = 10.27GLSWQADD389 pKa = 3.54LMKK392 pKa = 10.03WYY394 pKa = 10.94VDD396 pKa = 3.4DD397 pKa = 3.83TLVYY401 pKa = 10.09QVATPAEE408 pKa = 4.7LKK410 pKa = 10.34QPMYY414 pKa = 10.17MLVSLLTGTANSWQGLPTAANPTGTLQVDD443 pKa = 3.56YY444 pKa = 11.2VRR446 pKa = 11.84AYY448 pKa = 10.48QSTAVNVDD456 pKa = 3.32DD457 pKa = 5.53AGDD460 pKa = 3.45VAALGGHH467 pKa = 5.08YY468 pKa = 8.84TRR470 pKa = 11.84NADD473 pKa = 4.37GSDD476 pKa = 3.67LYY478 pKa = 11.44DD479 pKa = 3.78FTDD482 pKa = 3.54SAAMLVMDD490 pKa = 5.46ASGLSSGLHH499 pKa = 5.34GVLAGAAGSQVRR511 pKa = 11.84GSAGPMSFTGGAGVDD526 pKa = 3.43SFFFGVGNATVSGGDD541 pKa = 3.62GNDD544 pKa = 3.19SFVLTRR550 pKa = 11.84GVIAAGDD557 pKa = 3.89VIGDD561 pKa = 3.54FHH563 pKa = 8.55LDD565 pKa = 3.29LGNGTEE571 pKa = 4.27HH572 pKa = 7.5DD573 pKa = 3.97QLQLAGFSGDD583 pKa = 2.95AHH585 pKa = 7.76LDD587 pKa = 3.59VVSASGSTQIYY598 pKa = 9.08RR599 pKa = 11.84VVDD602 pKa = 3.66GTYY605 pKa = 9.86VSPNITIAVVNGAAHH620 pKa = 7.35LSAADD625 pKa = 3.6YY626 pKa = 11.57AFVV629 pKa = 3.54
Molecular weight: 64.77 kDa
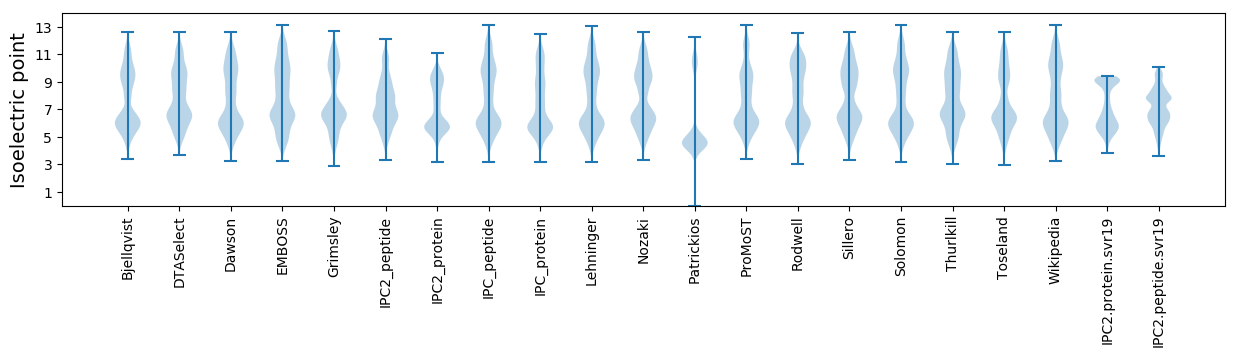
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4V2WLY4|A0A4V2WLY4_9PROT Corrinoid adenosyltransferase OS=Paracraurococcus sp. NE82 OX=2527868 GN=EXY23_04115 PE=3 SV=1
MM1 pKa = 7.36SGSARR6 pKa = 11.84ARR8 pKa = 11.84QPPKK12 pKa = 9.93PRR14 pKa = 11.84PAPRR18 pKa = 11.84AAPKK22 pKa = 10.01SAPRR26 pKa = 11.84QPARR30 pKa = 11.84AAPPALSGWRR40 pKa = 11.84RR41 pKa = 11.84WRR43 pKa = 11.84WVAVLVLAAVLGLPLAHH60 pKa = 7.0AMFGEE65 pKa = 4.9VVALLGGTFLLGLLVGRR82 pKa = 11.84WTARR86 pKa = 3.15
MM1 pKa = 7.36SGSARR6 pKa = 11.84ARR8 pKa = 11.84QPPKK12 pKa = 9.93PRR14 pKa = 11.84PAPRR18 pKa = 11.84AAPKK22 pKa = 10.01SAPRR26 pKa = 11.84QPARR30 pKa = 11.84AAPPALSGWRR40 pKa = 11.84RR41 pKa = 11.84WRR43 pKa = 11.84WVAVLVLAAVLGLPLAHH60 pKa = 7.0AMFGEE65 pKa = 4.9VVALLGGTFLLGLLVGRR82 pKa = 11.84WTARR86 pKa = 3.15
Molecular weight: 9.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1709467 |
25 |
2561 |
319.0 |
34.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
15.185 ± 0.054 | 0.881 ± 0.01 |
4.921 ± 0.027 | 5.74 ± 0.031 |
3.119 ± 0.02 | 9.542 ± 0.036 |
2.022 ± 0.016 | 3.806 ± 0.022 |
1.845 ± 0.021 | 11.158 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.229 ± 0.015 | 1.869 ± 0.017 |
6.445 ± 0.036 | 2.975 ± 0.02 |
8.783 ± 0.032 | 4.096 ± 0.021 |
4.836 ± 0.021 | 7.306 ± 0.03 |
1.52 ± 0.015 | 1.723 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
