
Tortoise microvirus 105
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
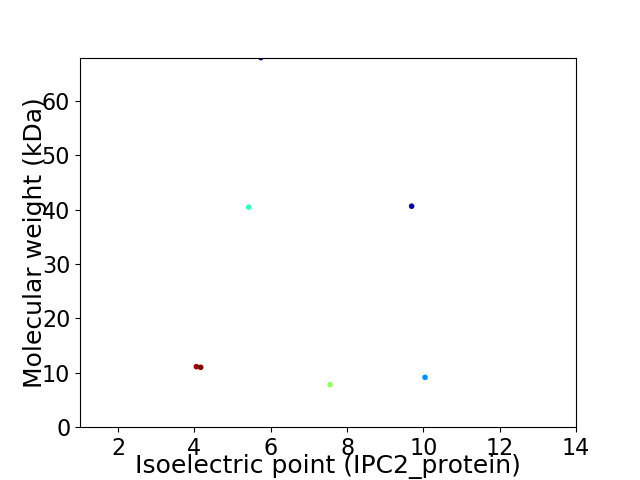
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
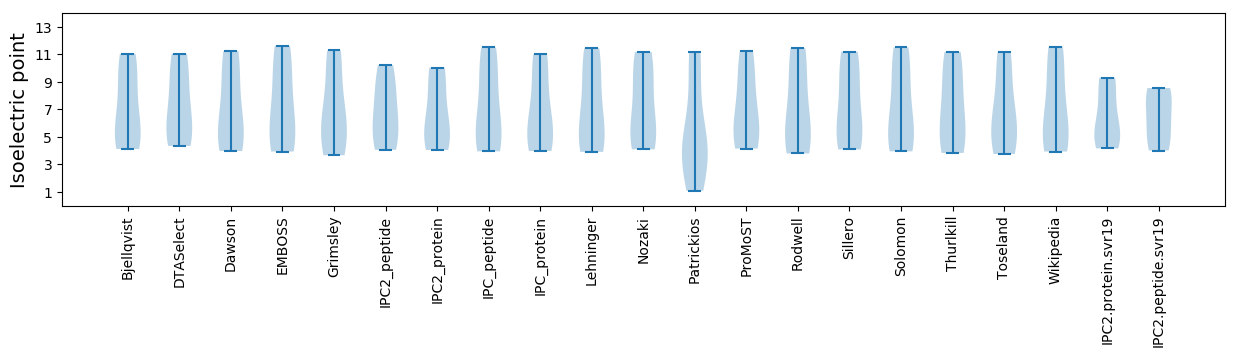
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W817|A0A4P8W817_9VIRU Replication initiation protein OS=Tortoise microvirus 105 OX=2583105 PE=4 SV=1
MM1 pKa = 6.74KK2 pKa = 10.32QIVIRR7 pKa = 11.84LIAPQVPSYY16 pKa = 11.0DD17 pKa = 3.47VTVGEE22 pKa = 4.91IIDD25 pKa = 3.94GQFAPVNDD33 pKa = 4.02INQIDD38 pKa = 4.15PKK40 pKa = 10.21LANVLIDD47 pKa = 5.57DD48 pKa = 4.24VTQSDD53 pKa = 3.75ILGGSSYY60 pKa = 10.43IRR62 pKa = 11.84SKK64 pKa = 11.13DD65 pKa = 3.47LSHH68 pKa = 7.46LLGSLYY74 pKa = 10.52ASNMISNVTMFPNFTVIDD92 pKa = 4.83LINYY96 pKa = 8.37EE97 pKa = 4.34SEE99 pKa = 4.19EE100 pKa = 4.23SS101 pKa = 3.4
MM1 pKa = 6.74KK2 pKa = 10.32QIVIRR7 pKa = 11.84LIAPQVPSYY16 pKa = 11.0DD17 pKa = 3.47VTVGEE22 pKa = 4.91IIDD25 pKa = 3.94GQFAPVNDD33 pKa = 4.02INQIDD38 pKa = 4.15PKK40 pKa = 10.21LANVLIDD47 pKa = 5.57DD48 pKa = 4.24VTQSDD53 pKa = 3.75ILGGSSYY60 pKa = 10.43IRR62 pKa = 11.84SKK64 pKa = 11.13DD65 pKa = 3.47LSHH68 pKa = 7.46LLGSLYY74 pKa = 10.52ASNMISNVTMFPNFTVIDD92 pKa = 4.83LINYY96 pKa = 8.37EE97 pKa = 4.34SEE99 pKa = 4.19EE100 pKa = 4.23SS101 pKa = 3.4
Molecular weight: 11.13 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8WAB0|A0A4P8WAB0_9VIRU Uncharacterized protein OS=Tortoise microvirus 105 OX=2583105 PE=4 SV=1
MM1 pKa = 7.47ACSHH5 pKa = 7.13PIWIRR10 pKa = 11.84NRR12 pKa = 11.84RR13 pKa = 11.84YY14 pKa = 9.64YY15 pKa = 10.65DD16 pKa = 3.04KK17 pKa = 11.0KK18 pKa = 10.02NPSRR22 pKa = 11.84LGSDD26 pKa = 3.14IQQSALALRR35 pKa = 11.84PWDD38 pKa = 4.19VSRR41 pKa = 11.84QWLMVPCGHH50 pKa = 7.23CDD52 pKa = 3.18DD53 pKa = 4.21CLRR56 pKa = 11.84RR57 pKa = 11.84LRR59 pKa = 11.84NDD61 pKa = 2.4WFVRR65 pKa = 11.84LDD67 pKa = 3.87RR68 pKa = 11.84EE69 pKa = 4.07LAHH72 pKa = 6.49NKK74 pKa = 9.88SINSQAVFITITISPKK90 pKa = 9.13YY91 pKa = 8.64YY92 pKa = 10.39DD93 pKa = 3.5RR94 pKa = 11.84AFSEE98 pKa = 4.41PAWFIRR104 pKa = 11.84KK105 pKa = 7.35WLEE108 pKa = 3.76RR109 pKa = 11.84VRR111 pKa = 11.84HH112 pKa = 4.97KK113 pKa = 10.74VGHH116 pKa = 5.22SFKK119 pKa = 10.69HH120 pKa = 6.15AFFQEE125 pKa = 4.74FGSHH129 pKa = 7.28PEE131 pKa = 4.05TGSEE135 pKa = 3.99PRR137 pKa = 11.84LHH139 pKa = 6.05FHH141 pKa = 6.84GFLFGLDD148 pKa = 3.0ISYY151 pKa = 11.1DD152 pKa = 3.33RR153 pKa = 11.84LRR155 pKa = 11.84AVVGDD160 pKa = 4.83LGFIWVARR168 pKa = 11.84ATQRR172 pKa = 11.84RR173 pKa = 11.84ARR175 pKa = 11.84YY176 pKa = 5.2TVKK179 pKa = 10.85YY180 pKa = 9.67VVKK183 pKa = 10.49QINYY187 pKa = 9.97DD188 pKa = 3.53GNNPKK193 pKa = 10.01LRR195 pKa = 11.84ALLQHH200 pKa = 6.37RR201 pKa = 11.84RR202 pKa = 11.84YY203 pKa = 7.92TRR205 pKa = 11.84KK206 pKa = 9.33FVSAGLGDD214 pKa = 3.79YY215 pKa = 10.6LGNKK219 pKa = 7.4PAPSFHH225 pKa = 6.08IRR227 pKa = 11.84SWSYY231 pKa = 10.81LDD233 pKa = 3.11TSSRR237 pKa = 11.84VNYY240 pKa = 10.12DD241 pKa = 3.17YY242 pKa = 11.23AIPRR246 pKa = 11.84YY247 pKa = 8.56YY248 pKa = 10.49NRR250 pKa = 11.84HH251 pKa = 6.08LKK253 pKa = 10.63PEE255 pKa = 4.32DD256 pKa = 3.36EE257 pKa = 4.46LRR259 pKa = 11.84RR260 pKa = 11.84SVLAADD266 pKa = 4.33TYY268 pKa = 11.94ARR270 pKa = 11.84FSKK273 pKa = 10.74SPLVRR278 pKa = 11.84HH279 pKa = 5.52IVSRR283 pKa = 11.84CVNLFIPEE291 pKa = 4.02ASLSRR296 pKa = 11.84RR297 pKa = 11.84ASYY300 pKa = 8.35TWQLEE305 pKa = 3.93QMNKK309 pKa = 8.88FRR311 pKa = 11.84TAGRR315 pKa = 11.84PLPAFDD321 pKa = 4.95PPTFLTRR328 pKa = 11.84QIVLFWYY335 pKa = 10.21DD336 pKa = 3.47NFKK339 pKa = 11.26LSITT343 pKa = 3.88
MM1 pKa = 7.47ACSHH5 pKa = 7.13PIWIRR10 pKa = 11.84NRR12 pKa = 11.84RR13 pKa = 11.84YY14 pKa = 9.64YY15 pKa = 10.65DD16 pKa = 3.04KK17 pKa = 11.0KK18 pKa = 10.02NPSRR22 pKa = 11.84LGSDD26 pKa = 3.14IQQSALALRR35 pKa = 11.84PWDD38 pKa = 4.19VSRR41 pKa = 11.84QWLMVPCGHH50 pKa = 7.23CDD52 pKa = 3.18DD53 pKa = 4.21CLRR56 pKa = 11.84RR57 pKa = 11.84LRR59 pKa = 11.84NDD61 pKa = 2.4WFVRR65 pKa = 11.84LDD67 pKa = 3.87RR68 pKa = 11.84EE69 pKa = 4.07LAHH72 pKa = 6.49NKK74 pKa = 9.88SINSQAVFITITISPKK90 pKa = 9.13YY91 pKa = 8.64YY92 pKa = 10.39DD93 pKa = 3.5RR94 pKa = 11.84AFSEE98 pKa = 4.41PAWFIRR104 pKa = 11.84KK105 pKa = 7.35WLEE108 pKa = 3.76RR109 pKa = 11.84VRR111 pKa = 11.84HH112 pKa = 4.97KK113 pKa = 10.74VGHH116 pKa = 5.22SFKK119 pKa = 10.69HH120 pKa = 6.15AFFQEE125 pKa = 4.74FGSHH129 pKa = 7.28PEE131 pKa = 4.05TGSEE135 pKa = 3.99PRR137 pKa = 11.84LHH139 pKa = 6.05FHH141 pKa = 6.84GFLFGLDD148 pKa = 3.0ISYY151 pKa = 11.1DD152 pKa = 3.33RR153 pKa = 11.84LRR155 pKa = 11.84AVVGDD160 pKa = 4.83LGFIWVARR168 pKa = 11.84ATQRR172 pKa = 11.84RR173 pKa = 11.84ARR175 pKa = 11.84YY176 pKa = 5.2TVKK179 pKa = 10.85YY180 pKa = 9.67VVKK183 pKa = 10.49QINYY187 pKa = 9.97DD188 pKa = 3.53GNNPKK193 pKa = 10.01LRR195 pKa = 11.84ALLQHH200 pKa = 6.37RR201 pKa = 11.84RR202 pKa = 11.84YY203 pKa = 7.92TRR205 pKa = 11.84KK206 pKa = 9.33FVSAGLGDD214 pKa = 3.79YY215 pKa = 10.6LGNKK219 pKa = 7.4PAPSFHH225 pKa = 6.08IRR227 pKa = 11.84SWSYY231 pKa = 10.81LDD233 pKa = 3.11TSSRR237 pKa = 11.84VNYY240 pKa = 10.12DD241 pKa = 3.17YY242 pKa = 11.23AIPRR246 pKa = 11.84YY247 pKa = 8.56YY248 pKa = 10.49NRR250 pKa = 11.84HH251 pKa = 6.08LKK253 pKa = 10.63PEE255 pKa = 4.32DD256 pKa = 3.36EE257 pKa = 4.46LRR259 pKa = 11.84RR260 pKa = 11.84SVLAADD266 pKa = 4.33TYY268 pKa = 11.94ARR270 pKa = 11.84FSKK273 pKa = 10.74SPLVRR278 pKa = 11.84HH279 pKa = 5.52IVSRR283 pKa = 11.84CVNLFIPEE291 pKa = 4.02ASLSRR296 pKa = 11.84RR297 pKa = 11.84ASYY300 pKa = 8.35TWQLEE305 pKa = 3.93QMNKK309 pKa = 8.88FRR311 pKa = 11.84TAGRR315 pKa = 11.84PLPAFDD321 pKa = 4.95PPTFLTRR328 pKa = 11.84QIVLFWYY335 pKa = 10.21DD336 pKa = 3.47NFKK339 pKa = 11.26LSITT343 pKa = 3.88
Molecular weight: 40.64 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1666 |
68 |
601 |
238.0 |
26.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.983 ± 1.423 | 0.72 ± 0.383 |
5.882 ± 0.818 | 3.842 ± 0.742 |
4.982 ± 0.814 | 6.242 ± 0.747 |
1.861 ± 0.57 | 5.522 ± 1.168 |
3.962 ± 1.034 | 7.623 ± 0.527 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.921 ± 0.447 | 5.642 ± 0.599 |
4.742 ± 0.729 | 4.682 ± 0.775 |
6.423 ± 1.342 | 8.764 ± 0.859 |
6.182 ± 1.155 | 5.942 ± 0.457 |
1.861 ± 0.371 | 5.222 ± 0.967 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
